| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
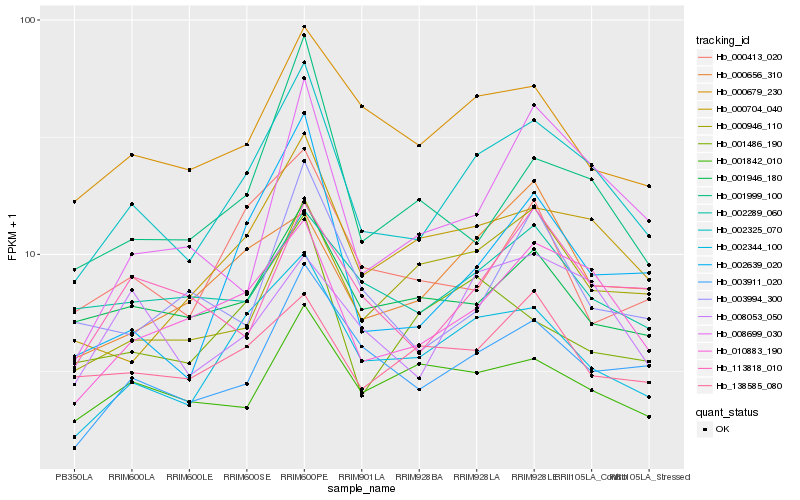
Hb_002325_070 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_010883_190 |
0.1068032267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002344_100 |
0.1068649053 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 4 |
Hb_003911_020 |
0.1258593266 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 5 |
Hb_000704_040 |
0.1313481646 |
- |
- |
aquaporin sip2.1, putative [Ricinus communis] |
| 6 |
Hb_002639_020 |
0.1369417339 |
- |
- |
PREDICTED: probable alpha-amylase 2 [Jatropha curcas] |
| 7 |
Hb_000679_230 |
0.1390487329 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 8 |
Hb_008053_050 |
0.1431558599 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001946_180 |
0.143999984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000946_110 |
0.1521238107 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 11 |
Hb_002289_060 |
0.1534207974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001999_100 |
0.1543055209 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 13 |
Hb_001842_010 |
0.1546020492 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_001486_190 |
0.156538741 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 15 |
Hb_000656_310 |
0.1573590794 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 16 |
Hb_000413_020 |
0.1576581176 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 17 |
Hb_113818_010 |
0.1580130278 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 18 |
Hb_008699_030 |
0.160681301 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 19 |
Hb_138585_080 |
0.1614440486 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 20 |
Hb_003994_300 |
0.1621843234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |