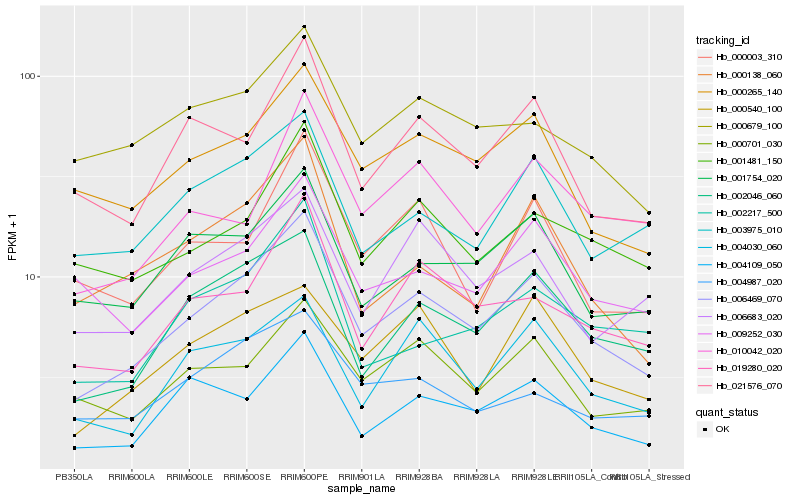
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006469_070 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 2 |
Hb_002046_060 |
0.0962762329 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 3 |
Hb_019280_020 |
0.1100980536 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000265_140 |
0.1101657277 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 5 |
Hb_004109_050 |
0.1108499494 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 6 |
Hb_021576_070 |
0.1143910777 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 7 |
Hb_001754_020 |
0.117323447 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 8 |
Hb_001481_150 |
0.1180120271 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 9 |
Hb_003975_010 |
0.1192683009 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 10 |
Hb_004030_060 |
0.1208509627 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 11 |
Hb_000540_100 |
0.1209684507 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 12 |
Hb_000701_030 |
0.1211351431 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 13 |
Hb_006683_020 |
0.1219285944 |
- |
- |
PREDICTED: nucleosome assembly protein 1;4 [Jatropha curcas] |
| 14 |
Hb_000138_060 |
0.1245140502 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 15 |
Hb_000679_100 |
0.1253765786 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010042_020 |
0.1268032459 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 17 |
Hb_009252_030 |
0.1277918269 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 18 |
Hb_002217_500 |
0.1286288486 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 19 |
Hb_000003_310 |
0.1295804049 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 20 |
Hb_004987_020 |
0.1296912986 |
- |
- |
- |