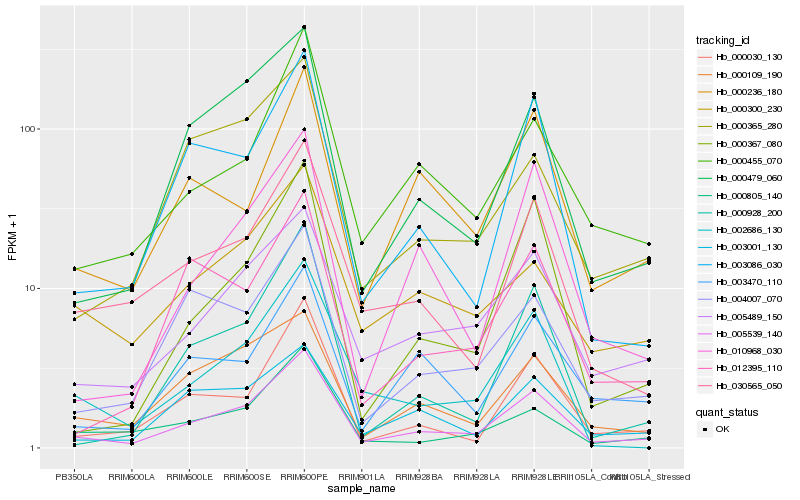
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_140 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 2 |
Hb_005489_150 |
0.1161032688 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 3 |
Hb_000479_060 |
0.1328762951 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 4 |
Hb_030565_050 |
0.1357807315 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 5 |
Hb_000455_070 |
0.1358900126 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 6 |
Hb_000030_130 |
0.1373560309 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000109_190 |
0.145086666 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 8 |
Hb_003086_030 |
0.146118678 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 9 |
Hb_000805_140 |
0.1473892667 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 10 |
Hb_004007_070 |
0.1474501057 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 11 |
Hb_002686_130 |
0.1513325177 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105650401 [Jatropha curcas] |
| 12 |
Hb_000367_080 |
0.1513597695 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 13 |
Hb_003470_110 |
0.1521011059 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000236_180 |
0.1530179937 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 15 |
Hb_003001_130 |
0.1537258984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010968_030 |
0.1540272629 |
- |
- |
PREDICTED: uncharacterized protein LOC105643149 [Jatropha curcas] |
| 17 |
Hb_000300_230 |
0.1547046299 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 18 |
Hb_000928_200 |
0.1591628897 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 19 |
Hb_012395_110 |
0.1599232118 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 20 |
Hb_000365_280 |
0.1612138385 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |