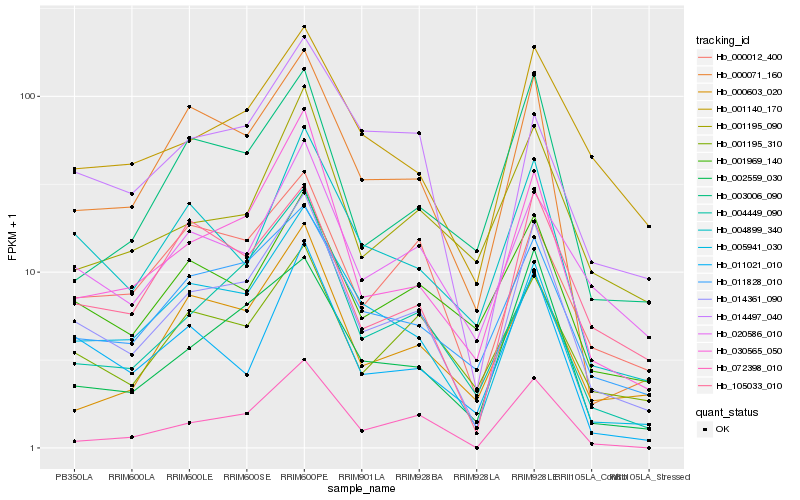
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014361_090 |
0.0 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 2 |
Hb_030565_050 |
0.115317864 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 3 |
Hb_011828_010 |
0.1224290374 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 4 |
Hb_002559_030 |
0.1240366733 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 5 |
Hb_005941_030 |
0.1250438062 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000071_160 |
0.1251707677 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 7 |
Hb_004899_340 |
0.1336418893 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_001195_310 |
0.1349288167 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_072398_010 |
0.135287205 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 10 |
Hb_020586_010 |
0.1366265236 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_011021_010 |
0.1386958819 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_001969_140 |
0.1388209416 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 13 |
Hb_001140_170 |
0.1397701352 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 14 |
Hb_105033_010 |
0.141856107 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001195_090 |
0.1479790597 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 16 |
Hb_000603_020 |
0.1494839737 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003006_090 |
0.1509063472 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 18 |
Hb_000012_400 |
0.1526039314 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 19 |
Hb_014497_040 |
0.1547396381 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004449_090 |
0.1571244629 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |