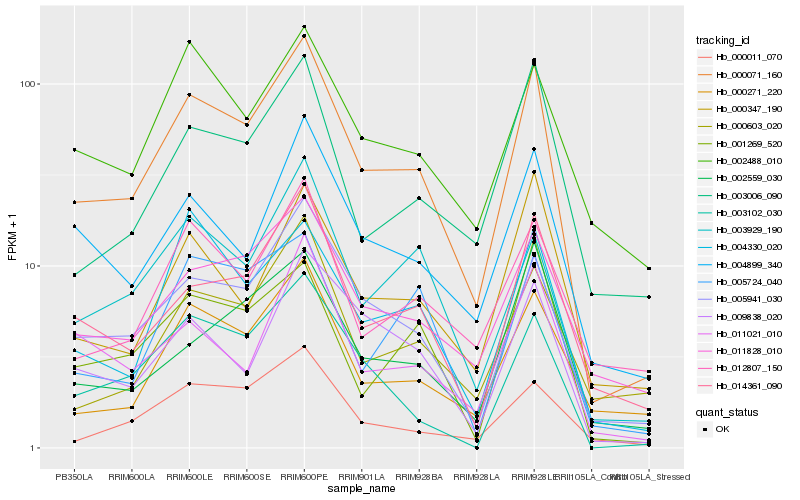
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000071_160 |
0.0 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 2 |
Hb_005941_030 |
0.1082182926 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_014361_090 |
0.1251707677 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 4 |
Hb_011828_010 |
0.1340899933 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 5 |
Hb_000271_220 |
0.1375428271 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 6 |
Hb_004899_340 |
0.1395722487 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 7 |
Hb_003929_190 |
0.1405422368 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 8 |
Hb_003006_090 |
0.1436377519 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 9 |
Hb_003102_030 |
0.1443037743 |
- |
- |
PREDICTED: uncharacterized protein LOC105644418 [Jatropha curcas] |
| 10 |
Hb_004330_020 |
0.1467431738 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 11 |
Hb_000011_070 |
0.147873171 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 12 |
Hb_001269_520 |
0.1483553358 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 13 |
Hb_009838_020 |
0.1487554749 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |
| 14 |
Hb_000347_190 |
0.1506049753 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 15 |
Hb_000603_020 |
0.1527198118 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002488_010 |
0.1535576981 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 17 |
Hb_012807_150 |
0.1535620231 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 18 |
Hb_011021_010 |
0.154170078 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_005724_040 |
0.1552926241 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 20 |
Hb_002559_030 |
0.156261079 |
- |
- |
oxidoreductase, putative [Ricinus communis] |