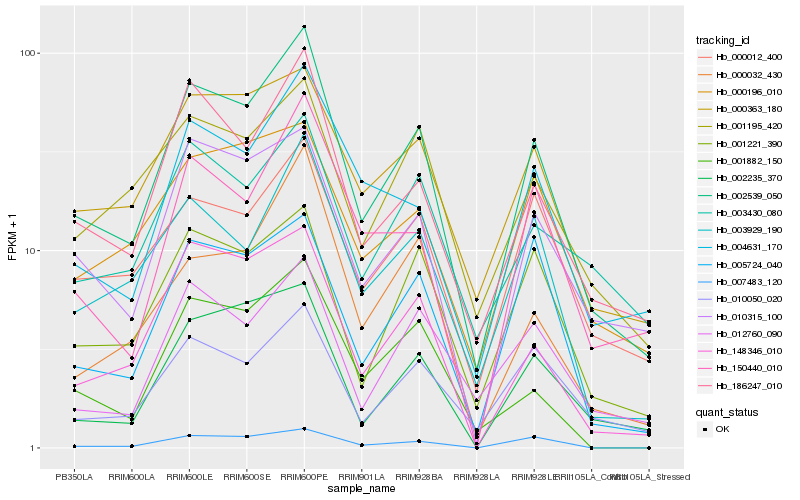
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002539_050 |
0.0 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 2 |
Hb_186247_010 |
0.1032117499 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 3 |
Hb_003929_190 |
0.1212443978 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 4 |
Hb_000363_180 |
0.1255421795 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 5 |
Hb_010315_100 |
0.1272484281 |
- |
- |
PREDICTED: uncharacterized protein LOC104420162 [Eucalyptus grandis] |
| 6 |
Hb_148346_010 |
0.1273064265 |
- |
- |
- |
| 7 |
Hb_004631_170 |
0.129111963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001221_390 |
0.1310939115 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 9 |
Hb_010050_020 |
0.1330206329 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 10 |
Hb_000012_400 |
0.1332283985 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 11 |
Hb_150440_010 |
0.13752703 |
- |
- |
- |
| 12 |
Hb_001195_420 |
0.1385734254 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 13 |
Hb_012760_090 |
0.1388838319 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 14 |
Hb_002235_370 |
0.1391979192 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 15 |
Hb_005724_040 |
0.1421212399 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 16 |
Hb_000032_430 |
0.1442508222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003430_080 |
0.1443267464 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_001882_150 |
0.1452602596 |
transcription factor |
TF Family: mTERF |
- |
| 19 |
Hb_007483_120 |
0.1464933915 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 20 |
Hb_000196_010 |
0.1466448119 |
- |
- |
Patellin-5, putative [Ricinus communis] |