| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
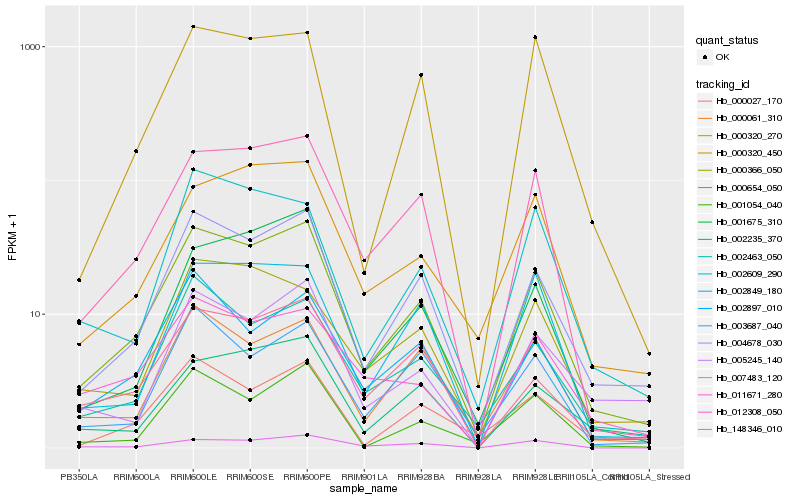
Hb_000654_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 2 |
Hb_004678_030 |
0.069455009 |
- |
- |
fructokinase [Manihot esculenta] |
| 3 |
Hb_012308_050 |
0.103133365 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 4 |
Hb_002463_050 |
0.1246636687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000320_270 |
0.1308489926 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000061_310 |
0.131905235 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001054_040 |
0.1349797387 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 8 |
Hb_148346_010 |
0.1358597486 |
- |
- |
- |
| 9 |
Hb_001675_310 |
0.1372625926 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002897_010 |
0.1380837314 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 11 |
Hb_000027_170 |
0.1382241444 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 12 |
Hb_007483_120 |
0.1386116073 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 13 |
Hb_002235_370 |
0.1400874689 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 14 |
Hb_000320_450 |
0.1416619053 |
- |
- |
secretory peroxidase [Hevea brasiliensis] |
| 15 |
Hb_011671_280 |
0.1442630574 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF106-like [Populus euphratica] |
| 16 |
Hb_005245_140 |
0.1443125569 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 17 |
Hb_002849_180 |
0.1444991275 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 18 |
Hb_002609_290 |
0.1483411398 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 19 |
Hb_003687_040 |
0.1488510878 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 20 |
Hb_000366_050 |
0.149574645 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |