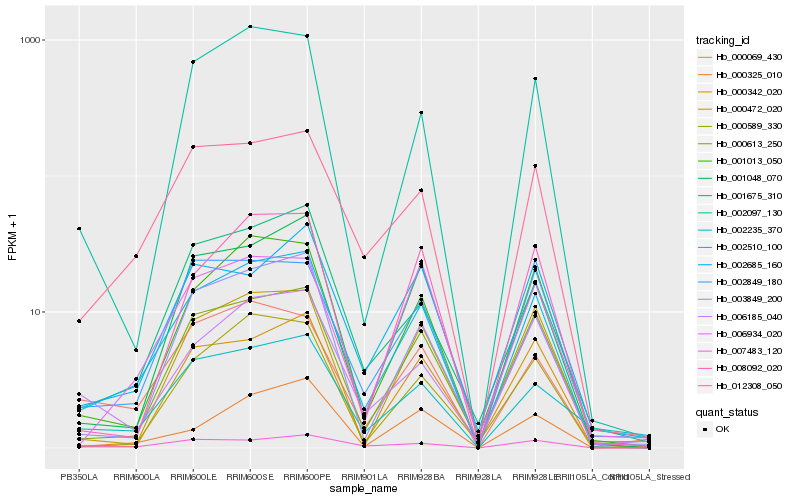
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002510_100 |
0.0 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 2 |
Hb_012308_050 |
0.1013528179 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 3 |
Hb_000613_250 |
0.1075413651 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 4 |
Hb_001013_050 |
0.1105469913 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_003849_200 |
0.1231416575 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 6 |
Hb_001048_070 |
0.1233773182 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 7 |
Hb_007483_120 |
0.1297721783 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 8 |
Hb_001675_310 |
0.1320018937 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002849_180 |
0.133260405 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 10 |
Hb_000069_430 |
0.1347721392 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 11 |
Hb_002097_130 |
0.13750568 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 12 |
Hb_000472_020 |
0.1376258524 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 13 |
Hb_008092_020 |
0.1390134303 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_006185_040 |
0.1394908798 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 15 |
Hb_000589_330 |
0.1411456194 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_000342_020 |
0.143203567 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 17 |
Hb_000325_010 |
0.1447853409 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 18 |
Hb_006934_020 |
0.1454346331 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 19 |
Hb_002235_370 |
0.1462095134 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 20 |
Hb_002685_160 |
0.1470099573 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |