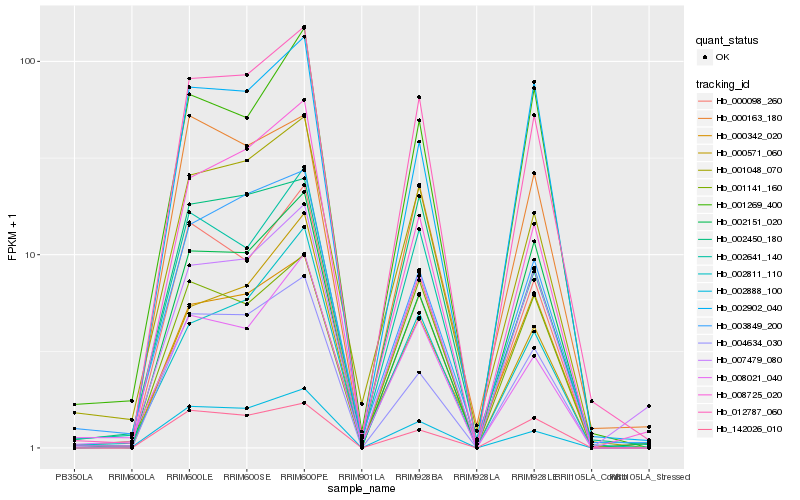
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012787_060 |
0.0 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 2 |
Hb_001048_070 |
0.0764503305 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 3 |
Hb_002888_100 |
0.0874711983 |
- |
- |
PREDICTED: uncharacterized protein LOC103699074, partial [Phoenix dactylifera] |
| 4 |
Hb_002641_140 |
0.0888208375 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 5 |
Hb_000098_260 |
0.0913129375 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 6 |
Hb_007479_080 |
0.1006328047 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004634_030 |
0.1012746074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008725_020 |
0.1016595898 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 9 |
Hb_000342_020 |
0.1028839873 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 10 |
Hb_003849_200 |
0.1047019857 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_000571_060 |
0.1071781583 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 12 |
Hb_001141_160 |
0.107551344 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 13 |
Hb_002902_040 |
0.1096699975 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 14 |
Hb_000163_180 |
0.109688887 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 15 |
Hb_142026_010 |
0.1125909401 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 16 |
Hb_001269_400 |
0.1145257973 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 17 |
Hb_002151_020 |
0.1149087817 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 18 |
Hb_002450_180 |
0.1152099594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002811_110 |
0.1152714642 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 20 |
Hb_008021_040 |
0.1157373701 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |