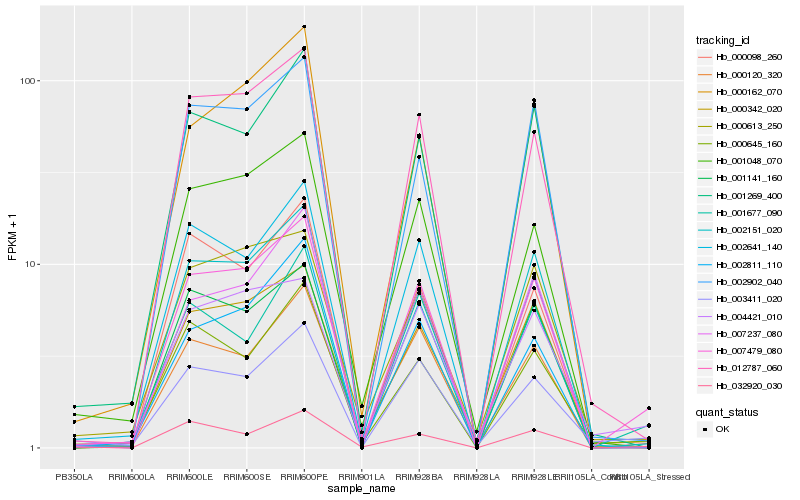
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007479_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 2 |
Hb_012787_060 |
0.1006328047 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 3 |
Hb_000342_020 |
0.1070631001 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 4 |
Hb_001269_400 |
0.1089696014 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 5 |
Hb_002151_020 |
0.1090936113 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 6 |
Hb_003411_020 |
0.1091862265 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 7 |
Hb_001048_070 |
0.1147707061 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 8 |
Hb_004421_010 |
0.115166097 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 9 |
Hb_000162_070 |
0.1173338004 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 10 |
Hb_001677_090 |
0.1183912104 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |
| 11 |
Hb_002902_040 |
0.1184704841 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 12 |
Hb_007237_080 |
0.1186795776 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000645_160 |
0.1187438348 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 14 |
Hb_000120_320 |
0.1201784502 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000613_250 |
0.1204179717 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 16 |
Hb_002811_110 |
0.123586293 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 17 |
Hb_000098_260 |
0.1250452818 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 18 |
Hb_002641_140 |
0.1262447281 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_001141_160 |
0.1264338777 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 20 |
Hb_032920_030 |
0.1266413224 |
- |
- |
signal transducer, putative [Ricinus communis] |