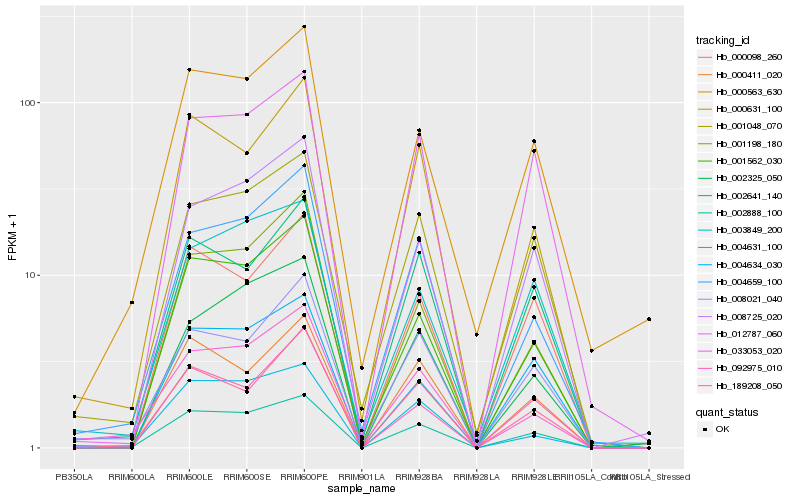
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002888_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103699074, partial [Phoenix dactylifera] |
| 2 |
Hb_001562_030 |
0.0800383556 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012787_060 |
0.0874711983 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 4 |
Hb_000098_260 |
0.095448964 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 5 |
Hb_001048_070 |
0.0989531898 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 6 |
Hb_008725_020 |
0.1004459299 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 7 |
Hb_004634_030 |
0.1013432117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000631_100 |
0.1053286119 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 9 |
Hb_000411_020 |
0.1054834179 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_004659_100 |
0.1058125029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002641_140 |
0.1058188302 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 12 |
Hb_001198_180 |
0.108573812 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 13 |
Hb_004631_100 |
0.1087456942 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 14 |
Hb_003849_200 |
0.1097493348 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 15 |
Hb_189208_050 |
0.1111522602 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000563_630 |
0.112241836 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 17 |
Hb_002325_050 |
0.1177208543 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 18 |
Hb_008021_040 |
0.1189747798 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 19 |
Hb_033053_020 |
0.1197062791 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 20 |
Hb_092975_010 |
0.120164159 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |