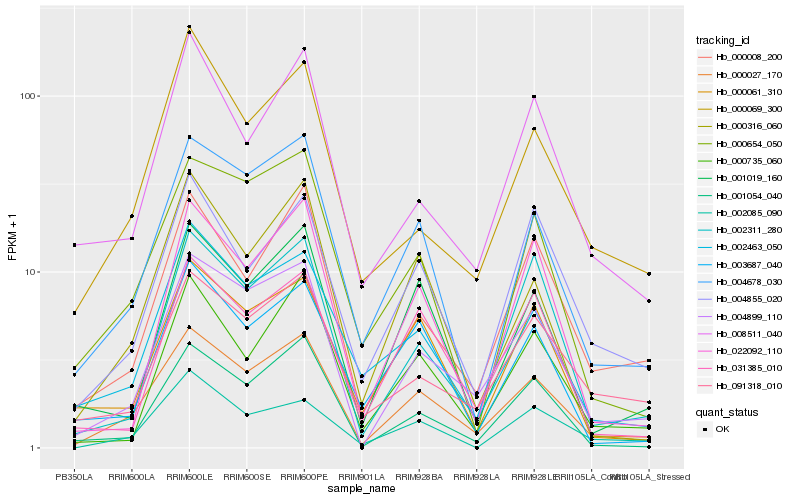
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_170 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 2 |
Hb_004899_110 |
0.0955364411 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 3 |
Hb_004678_030 |
0.1085542088 |
- |
- |
fructokinase [Manihot esculenta] |
| 4 |
Hb_000008_200 |
0.1281991744 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 5 |
Hb_001054_040 |
0.13468054 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 6 |
Hb_091318_010 |
0.1350646199 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 7 |
Hb_000316_060 |
0.135144897 |
- |
- |
hypothetical protein CISIN_1g017081mg [Citrus sinensis] |
| 8 |
Hb_002463_050 |
0.1378676858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000654_050 |
0.1382241444 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 10 |
Hb_001019_160 |
0.1383517639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004855_020 |
0.1389156989 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 12 |
Hb_000061_310 |
0.1468909113 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000735_060 |
0.148549455 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 14 |
Hb_002311_280 |
0.1495734559 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 15 |
Hb_000069_300 |
0.1500203025 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_031385_010 |
0.1509817599 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 17 |
Hb_008511_040 |
0.1528730939 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 18 |
Hb_022092_110 |
0.1540473844 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 19 |
Hb_003687_040 |
0.1541754782 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 20 |
Hb_002085_090 |
0.1549798442 |
- |
- |
- |