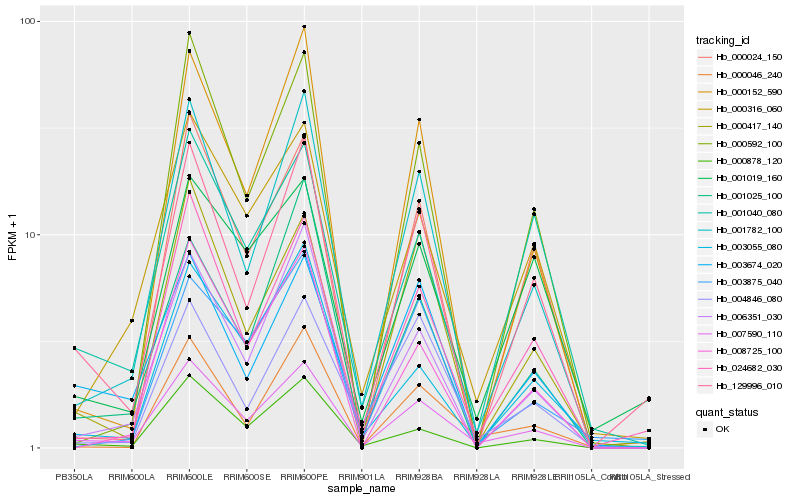
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007590_110 |
0.0 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 2 |
Hb_001782_100 |
0.1042972755 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_000316_060 |
0.1217352766 |
- |
- |
hypothetical protein CISIN_1g017081mg [Citrus sinensis] |
| 4 |
Hb_024682_030 |
0.1292690337 |
- |
- |
PREDICTED: DNA replication licensing factor MCM2 [Jatropha curcas] |
| 5 |
Hb_129996_010 |
0.1301688167 |
- |
- |
hypothetical protein PRUPE_ppa013547mg [Prunus persica] |
| 6 |
Hb_006351_030 |
0.1310594061 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MARCH8-like [Jatropha curcas] |
| 7 |
Hb_000417_140 |
0.1403047108 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 8 |
Hb_003674_020 |
0.1405199835 |
- |
- |
PREDICTED: DNA replication licensing factor MCM6 [Jatropha curcas] |
| 9 |
Hb_000878_120 |
0.1409902898 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 10 |
Hb_000152_590 |
0.1503623112 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 11 |
Hb_008725_100 |
0.1526952812 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 12 |
Hb_000592_100 |
0.1540585117 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 13 |
Hb_001040_080 |
0.1544800262 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 14 |
Hb_000024_150 |
0.1555494845 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 15 |
Hb_000046_240 |
0.1557973553 |
- |
- |
PREDICTED: uncharacterized protein LOC105631837 [Jatropha curcas] |
| 16 |
Hb_003055_080 |
0.1594886456 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004846_080 |
0.1635735587 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 18 |
Hb_001019_160 |
0.1648476989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001025_100 |
0.1653355377 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003875_040 |
0.1663884005 |
- |
- |
PREDICTED: endoglucanase 11-like [Jatropha curcas] |