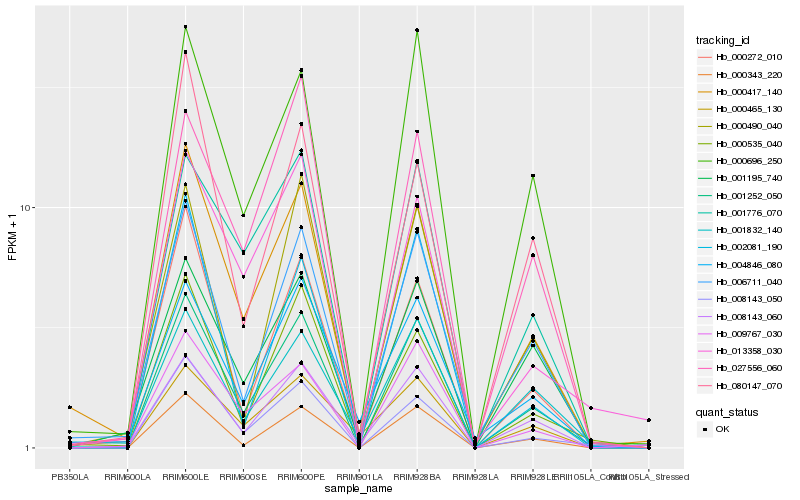
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001252_050 |
0.0 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 2 |
Hb_000343_220 |
0.0835976578 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 3 |
Hb_008143_050 |
0.098852953 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 4 |
Hb_000696_250 |
0.0990958842 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 5 |
Hb_006711_040 |
0.1066382697 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 6 |
Hb_000417_140 |
0.1105887849 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 7 |
Hb_008143_060 |
0.1119031152 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 8 |
Hb_009767_030 |
0.1149252767 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 9 |
Hb_001776_070 |
0.1177795413 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 10 |
Hb_001832_140 |
0.1185757054 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 11 |
Hb_027556_060 |
0.1191156958 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 12 |
Hb_000535_040 |
0.1193758648 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 13 |
Hb_000272_010 |
0.1291159532 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 14 |
Hb_001195_740 |
0.1313250055 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000490_040 |
0.136202432 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |
| 16 |
Hb_013358_030 |
0.1370428694 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 17 |
Hb_000465_130 |
0.1416425713 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0001s15550g [Populus trichocarpa] |
| 18 |
Hb_002081_190 |
0.1445379102 |
- |
- |
hypothetical protein EUGRSUZ_I010391, partial [Eucalyptus grandis] |
| 19 |
Hb_004846_080 |
0.147485761 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 20 |
Hb_080147_070 |
0.1490526499 |
- |
- |
Rop5 [Hevea brasiliensis] |