| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
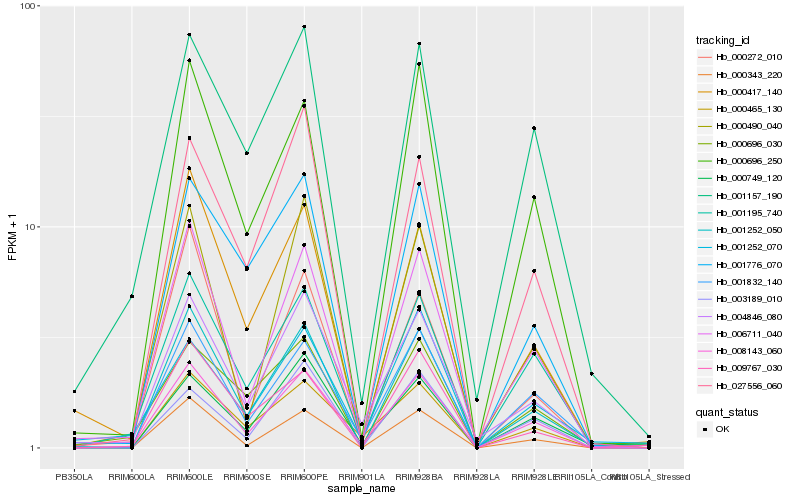
Hb_008143_060 |
0.0 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 2 |
Hb_001195_740 |
0.0856663632 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006711_040 |
0.099341318 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 4 |
Hb_001832_140 |
0.1003571671 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 5 |
Hb_001252_050 |
0.1119031152 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 6 |
Hb_000696_250 |
0.1147230215 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 7 |
Hb_000465_130 |
0.1204044648 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0001s15550g [Populus trichocarpa] |
| 8 |
Hb_000749_120 |
0.1206074802 |
- |
- |
PREDICTED: uncharacterized protein LOC105644255 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004846_080 |
0.1226376024 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 10 |
Hb_000343_220 |
0.1251864833 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 11 |
Hb_000490_040 |
0.131613046 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |
| 12 |
Hb_027556_060 |
0.135546203 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 13 |
Hb_009767_030 |
0.1357802674 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 14 |
Hb_000417_140 |
0.1381270137 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 15 |
Hb_000272_010 |
0.1406758709 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 16 |
Hb_001157_190 |
0.1421121599 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 17 |
Hb_001776_070 |
0.14585008 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 18 |
Hb_001252_070 |
0.1503078289 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 19 |
Hb_003189_010 |
0.1519581565 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 20 |
Hb_000696_030 |
0.1526365446 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |