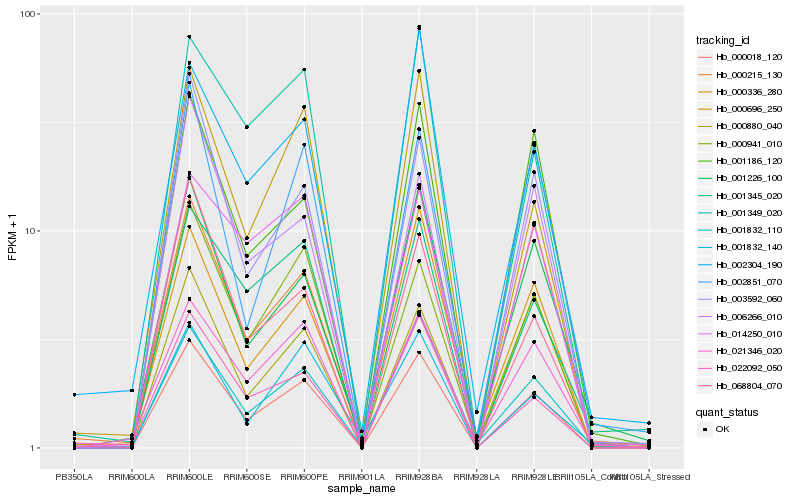
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000018_120 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000696_250 |
0.0990816276 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 3 |
Hb_003592_060 |
0.1056607271 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |
| 4 |
Hb_001832_110 |
0.1090922382 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000941_010 |
0.1095676169 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 6 |
Hb_000336_280 |
0.1171006138 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 7 |
Hb_022092_050 |
0.1183090931 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 8 |
Hb_001349_020 |
0.1188567739 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 9 |
Hb_021346_020 |
0.1216056137 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 10 |
Hb_068804_070 |
0.1229038205 |
- |
- |
hypothetical protein JCGZ_12627 [Jatropha curcas] |
| 11 |
Hb_001345_020 |
0.1234003778 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 12 |
Hb_001186_120 |
0.1249538874 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 13 |
Hb_001226_100 |
0.1252668788 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002851_070 |
0.1297745414 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 15 |
Hb_014250_010 |
0.1308148795 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 16 |
Hb_006266_010 |
0.1322150614 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000215_130 |
0.132818586 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 18 |
Hb_000880_040 |
0.1345861647 |
- |
- |
- |
| 19 |
Hb_002304_190 |
0.1354424744 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |
| 20 |
Hb_001832_140 |
0.1360944803 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |