| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
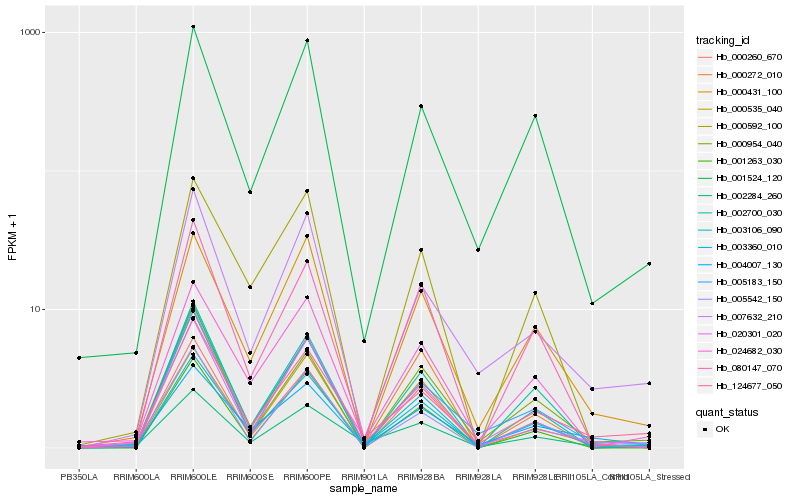
Hb_003360_010 |
0.0 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 2 |
Hb_024682_030 |
0.0817926 |
- |
- |
PREDICTED: DNA replication licensing factor MCM2 [Jatropha curcas] |
| 3 |
Hb_000260_670 |
0.083983672 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 4 |
Hb_000431_100 |
0.0891994846 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 5 |
Hb_124677_050 |
0.0907439218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001524_120 |
0.0922391803 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_005183_150 |
0.0956732135 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 8 |
Hb_080147_070 |
0.1032804907 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 9 |
Hb_002284_260 |
0.1105717862 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 10 |
Hb_020301_020 |
0.1112901374 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_002700_030 |
0.1151280069 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 12 |
Hb_007632_210 |
0.1162479251 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 13 |
Hb_004007_130 |
0.1168689806 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 14 |
Hb_000954_040 |
0.1179264509 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 15 |
Hb_000592_100 |
0.1196090201 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 16 |
Hb_005542_150 |
0.1198338074 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 17 |
Hb_000272_010 |
0.1225002243 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 18 |
Hb_001263_030 |
0.1225350449 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 19 |
Hb_003106_090 |
0.1258981666 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 20 |
Hb_000535_040 |
0.1273246822 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |