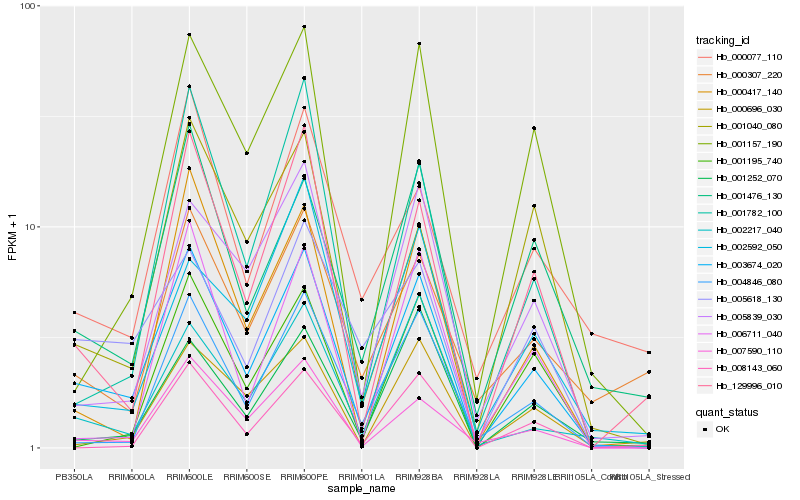
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003674_020 |
0.0 |
- |
- |
PREDICTED: DNA replication licensing factor MCM6 [Jatropha curcas] |
| 2 |
Hb_007590_110 |
0.1405199835 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 3 |
Hb_129996_010 |
0.1461651235 |
- |
- |
hypothetical protein PRUPE_ppa013547mg [Prunus persica] |
| 4 |
Hb_004846_080 |
0.1566421412 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 5 |
Hb_001476_130 |
0.1574594446 |
- |
- |
PREDICTED: probable protein S-acyltransferase 22 [Jatropha curcas] |
| 6 |
Hb_000696_030 |
0.1617696074 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001782_100 |
0.1620106966 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 8 |
Hb_000417_140 |
0.1626286462 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 9 |
Hb_005839_030 |
0.1633250777 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 10 |
Hb_001157_190 |
0.1650064865 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 11 |
Hb_002217_040 |
0.165314922 |
- |
- |
PREDICTED: boron transporter 1-like [Jatropha curcas] |
| 12 |
Hb_005618_130 |
0.167875199 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 13 |
Hb_006711_040 |
0.1746815554 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 14 |
Hb_001195_740 |
0.1757254033 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001252_070 |
0.1789172669 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 16 |
Hb_000077_110 |
0.1876822118 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 17 |
Hb_001040_080 |
0.1877553593 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 18 |
Hb_000307_220 |
0.190183909 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 19 |
Hb_008143_060 |
0.1907754625 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 20 |
Hb_002592_050 |
0.1925149147 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |