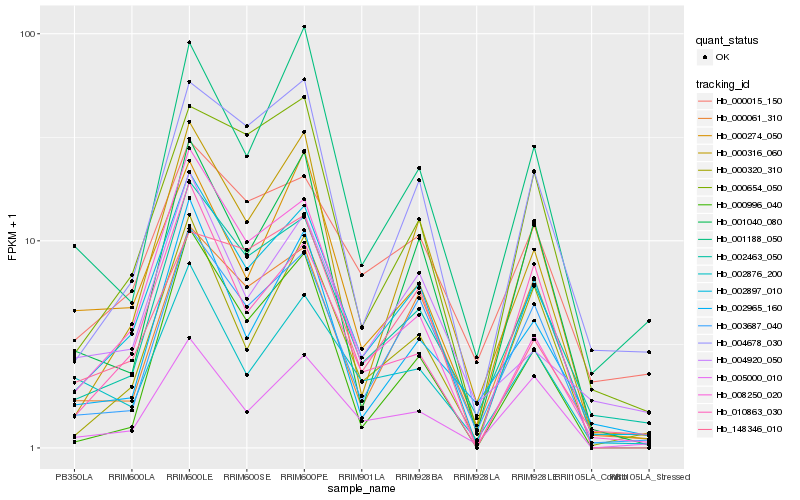
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002897_010 |
0.0 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 2 |
Hb_002463_050 |
0.0966236615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003687_040 |
0.1109852344 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 4 |
Hb_000316_060 |
0.1283505279 |
- |
- |
hypothetical protein CISIN_1g017081mg [Citrus sinensis] |
| 5 |
Hb_000320_310 |
0.1290617734 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 6 |
Hb_008250_020 |
0.1305382887 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004678_030 |
0.1321314392 |
- |
- |
fructokinase [Manihot esculenta] |
| 8 |
Hb_000061_310 |
0.1340286394 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001040_080 |
0.1373841805 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 10 |
Hb_005000_010 |
0.1379886969 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000654_050 |
0.1380837314 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 12 |
Hb_010863_030 |
0.1404689403 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 13 |
Hb_000015_150 |
0.1436924686 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 14 |
Hb_002876_200 |
0.146473211 |
- |
- |
PREDICTED: uncharacterized protein LOC105641209 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000274_050 |
0.146681517 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 16 |
Hb_002965_160 |
0.1551278102 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 17 |
Hb_004920_050 |
0.1554584242 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 18 |
Hb_001188_050 |
0.1566889677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000996_040 |
0.157176978 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 20 |
Hb_148346_010 |
0.1577769339 |
- |
- |
- |