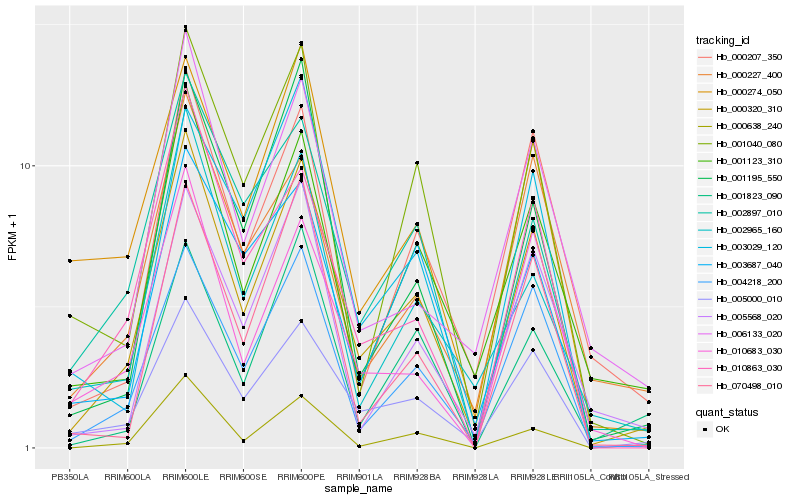
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_310 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 2 |
Hb_005000_010 |
0.0848071778 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004218_200 |
0.0997838407 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 4 |
Hb_010863_030 |
0.1288353789 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 5 |
Hb_002897_010 |
0.1290617734 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 6 |
Hb_001123_310 |
0.1369853654 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 7 |
Hb_001195_550 |
0.1391063069 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001040_080 |
0.1420614168 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 9 |
Hb_070498_010 |
0.1435786549 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 10 |
Hb_003687_040 |
0.1454763162 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 11 |
Hb_006133_020 |
0.1458617007 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 12 |
Hb_010683_030 |
0.1461482427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000207_350 |
0.1464511599 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 14 |
Hb_000274_050 |
0.1468808216 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_003029_120 |
0.1473357142 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 16 |
Hb_000227_400 |
0.1473990029 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 17 |
Hb_001823_090 |
0.1480767215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000638_240 |
0.1498910217 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Prunus mume] |
| 19 |
Hb_005568_020 |
0.1502475009 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002965_160 |
0.1505984215 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |