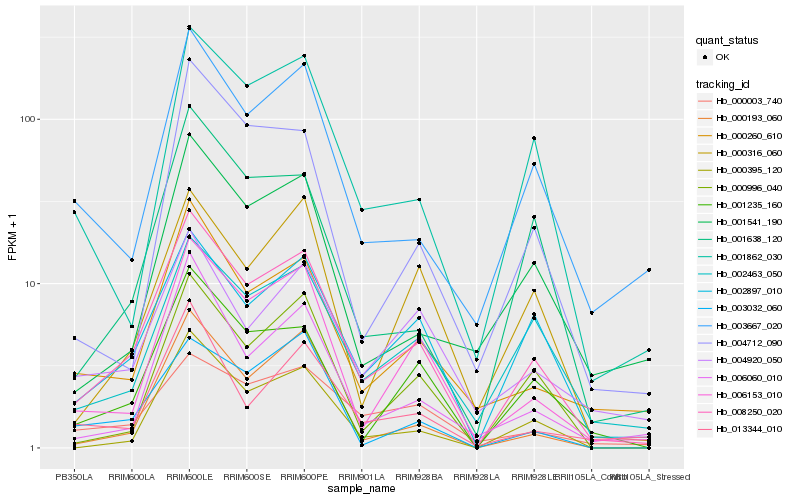
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008250_020 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_006153_010 |
0.1294917685 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 [Populus euphratica] |
| 3 |
Hb_002897_010 |
0.1305382887 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 4 |
Hb_001235_160 |
0.1362612729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000395_120 |
0.142379825 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 6 |
Hb_000260_610 |
0.1430758041 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 7 |
Hb_002463_050 |
0.1509238573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000193_060 |
0.1509406423 |
- |
- |
PREDICTED: F-box protein At4g35930 [Jatropha curcas] |
| 9 |
Hb_000996_040 |
0.1533855109 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 10 |
Hb_001862_030 |
0.159763983 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 11 |
Hb_006060_010 |
0.1598749853 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 12 |
Hb_001638_120 |
0.1599175366 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 13 |
Hb_003667_020 |
0.1601744664 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 14 |
Hb_000003_740 |
0.1700062242 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 15 |
Hb_003032_060 |
0.1704751205 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 16 |
Hb_004712_090 |
0.1716578684 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 17 |
Hb_000316_060 |
0.1728544183 |
- |
- |
hypothetical protein CISIN_1g017081mg [Citrus sinensis] |
| 18 |
Hb_001541_190 |
0.1734453864 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 19 |
Hb_004920_050 |
0.1741380844 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 20 |
Hb_013344_010 |
0.1754193846 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |