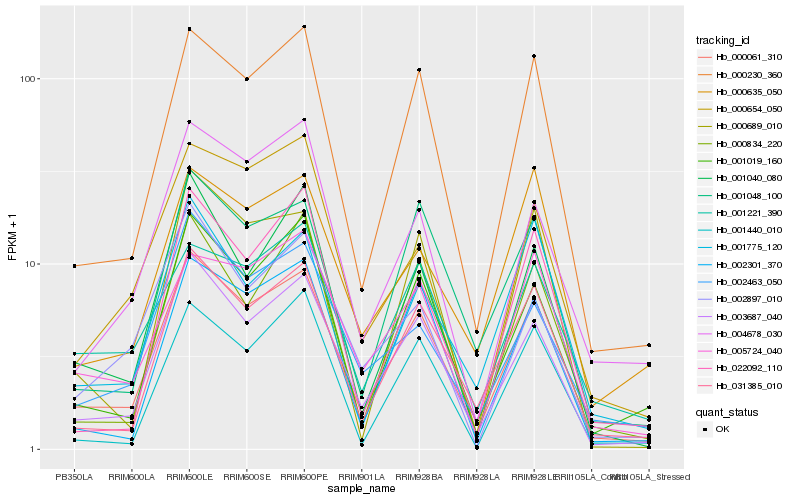
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_310 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003687_040 |
0.0607742234 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 3 |
Hb_000230_360 |
0.0725496675 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 4 |
Hb_031385_010 |
0.0841381792 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 5 |
Hb_001019_160 |
0.1053184784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001048_100 |
0.1088018161 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 7 |
Hb_002301_370 |
0.1149250894 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 8 |
Hb_001040_080 |
0.1154183836 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 9 |
Hb_005724_040 |
0.1202455424 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 10 |
Hb_004678_030 |
0.1228274448 |
- |
- |
fructokinase [Manihot esculenta] |
| 11 |
Hb_002463_050 |
0.1228559465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001775_120 |
0.1253112287 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 13 |
Hb_000834_220 |
0.1282685675 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 14 |
Hb_001221_390 |
0.1289327876 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 15 |
Hb_000654_050 |
0.131905235 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 16 |
Hb_000635_050 |
0.1331950949 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 17 |
Hb_000689_010 |
0.1336210364 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 18 |
Hb_002897_010 |
0.1340286394 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 19 |
Hb_022092_110 |
0.1342557963 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 20 |
Hb_001440_010 |
0.1353689353 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |