| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
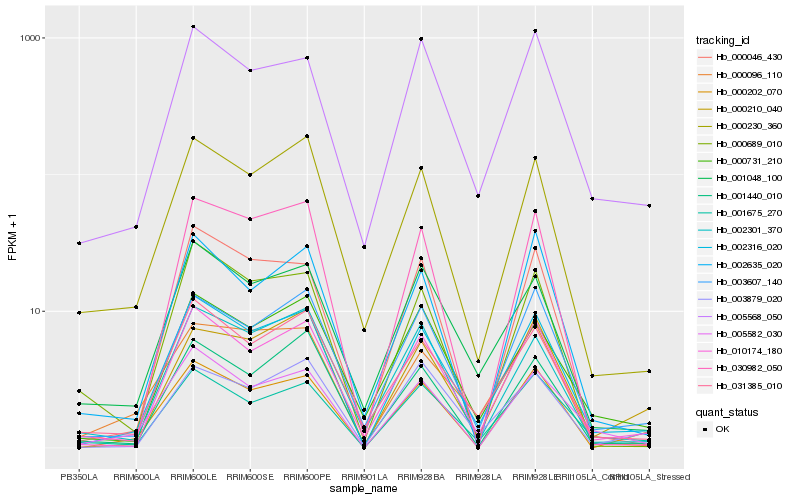
Hb_002316_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 2 |
Hb_010174_180 |
0.07529422 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_031385_010 |
0.0970108493 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 4 |
Hb_001675_270 |
0.0975660153 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 5 |
Hb_003607_140 |
0.1013357766 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 6 |
Hb_000230_360 |
0.1080390496 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 7 |
Hb_001048_100 |
0.1128658632 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 8 |
Hb_005582_030 |
0.1135930774 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A-like [Jatropha curcas] |
| 9 |
Hb_005568_050 |
0.1175107272 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 10 |
Hb_001440_010 |
0.1179018887 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 11 |
Hb_000731_210 |
0.1190934272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002301_370 |
0.1199503084 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 13 |
Hb_000202_070 |
0.1213055884 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 14 |
Hb_000046_430 |
0.1221627295 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 15 |
Hb_000096_110 |
0.1240819829 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 16 |
Hb_000210_040 |
0.125195965 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 17 |
Hb_030982_050 |
0.1253867098 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 18 |
Hb_003879_020 |
0.1282865491 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 19 |
Hb_000689_010 |
0.1310530416 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_002635_020 |
0.1317770894 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |