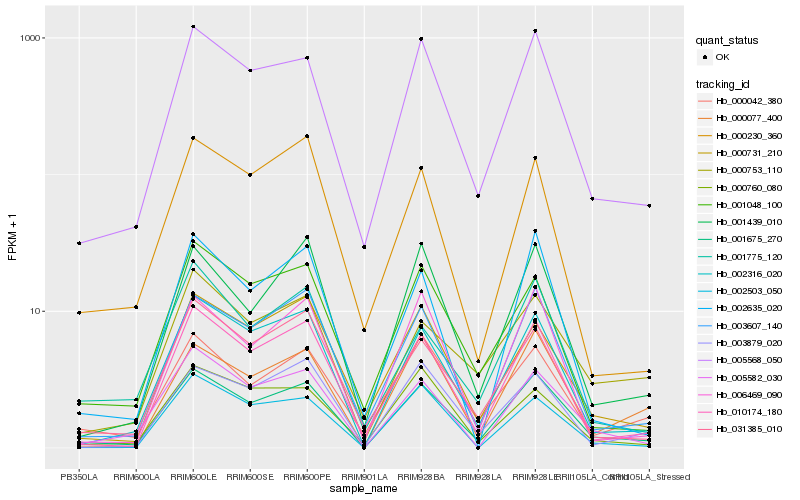
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001675_270 |
0.0 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 2 |
Hb_005568_050 |
0.0801035741 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 3 |
Hb_002316_020 |
0.0975660153 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 4 |
Hb_003607_140 |
0.105495868 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 5 |
Hb_010174_180 |
0.1072727233 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_005582_030 |
0.121977906 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A-like [Jatropha curcas] |
| 7 |
Hb_001439_010 |
0.1235699648 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 8 |
Hb_006469_090 |
0.1261797356 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 9 |
Hb_000077_400 |
0.1324851098 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000042_380 |
0.1328756427 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 11 |
Hb_002635_020 |
0.1347384736 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 12 |
Hb_000731_210 |
0.1375025641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_031385_010 |
0.137911324 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 14 |
Hb_001048_100 |
0.1395739641 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 15 |
Hb_003879_020 |
0.1421095147 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 16 |
Hb_000760_080 |
0.1435448302 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 17 |
Hb_001775_120 |
0.1438331633 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 18 |
Hb_000753_110 |
0.1440633264 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X6 [Jatropha curcas] |
| 19 |
Hb_002503_050 |
0.1456372181 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 20 |
Hb_000230_360 |
0.1460821133 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |