| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
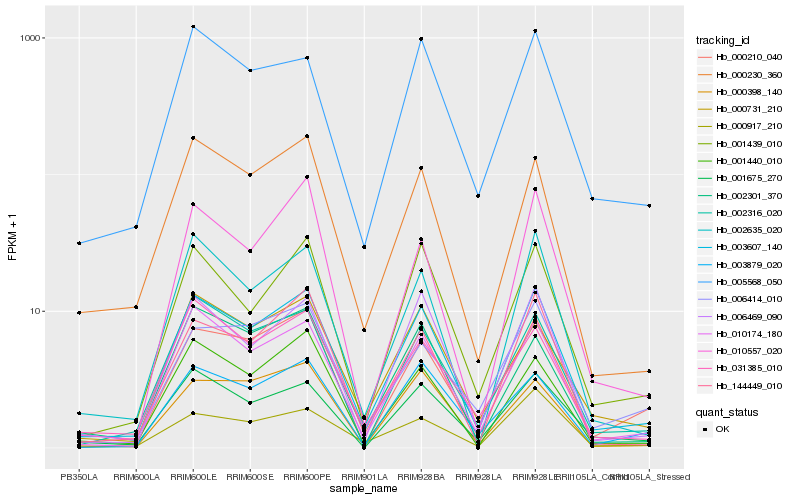
Hb_003607_140 |
0.0 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 2 |
Hb_010174_180 |
0.0756157517 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_006469_090 |
0.0947962222 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 4 |
Hb_001439_010 |
0.096231105 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 5 |
Hb_002635_020 |
0.1008438932 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 6 |
Hb_002316_020 |
0.1013357766 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 7 |
Hb_000210_040 |
0.1035653701 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 8 |
Hb_000230_360 |
0.1042070316 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 9 |
Hb_001675_270 |
0.105495868 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 10 |
Hb_144449_010 |
0.1099868474 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 11 |
Hb_001440_010 |
0.1113149411 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 12 |
Hb_005568_050 |
0.1125508394 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 13 |
Hb_003879_020 |
0.1140953942 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 14 |
Hb_000731_210 |
0.1153844451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_031385_010 |
0.1193066096 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 16 |
Hb_002301_370 |
0.1230781426 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 17 |
Hb_000398_140 |
0.1233888425 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 18 |
Hb_000917_210 |
0.1252166087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010557_020 |
0.1268980706 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 20 |
Hb_006414_010 |
0.1279489692 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |