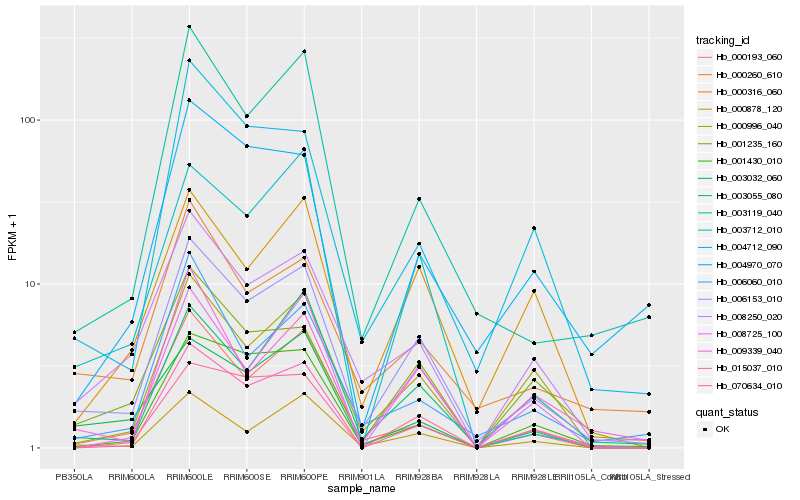
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006153_010 |
0.0 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 [Populus euphratica] |
| 2 |
Hb_008250_020 |
0.1294917685 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_003119_040 |
0.1383505018 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 4 |
Hb_000878_120 |
0.1414527459 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 5 |
Hb_000260_610 |
0.1432975215 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 6 |
Hb_000193_060 |
0.1462655715 |
- |
- |
PREDICTED: F-box protein At4g35930 [Jatropha curcas] |
| 7 |
Hb_015037_010 |
0.1484448642 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 8 |
Hb_000996_040 |
0.148610147 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 9 |
Hb_001235_160 |
0.153072377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000316_060 |
0.1542540699 |
- |
- |
hypothetical protein CISIN_1g017081mg [Citrus sinensis] |
| 11 |
Hb_009339_040 |
0.1545872906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001430_010 |
0.1574826581 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_006060_010 |
0.158208349 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 14 |
Hb_004712_090 |
0.1582787149 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 15 |
Hb_003055_080 |
0.1628285609 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003032_060 |
0.1629665729 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 17 |
Hb_070634_010 |
0.1663030651 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 18 |
Hb_003712_010 |
0.1679470626 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_004970_070 |
0.168606775 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 20 |
Hb_008725_100 |
0.1700121269 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |