| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
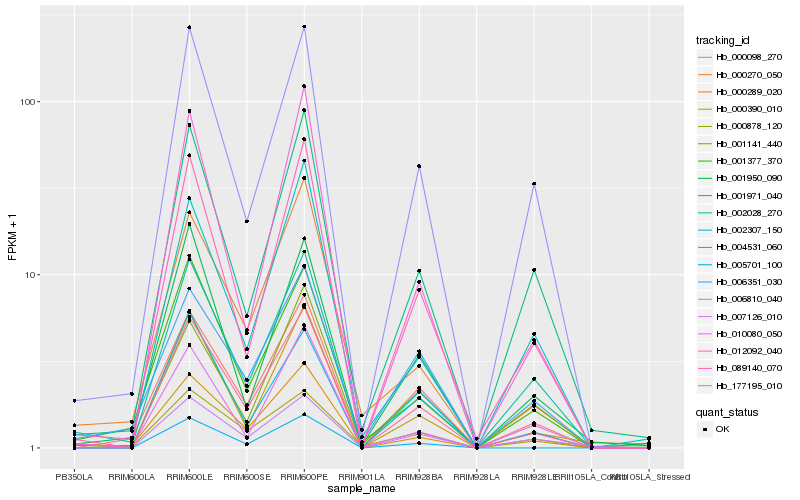
Hb_177195_010 |
0.0 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 3, partial [Theobroma cacao] |
| 2 |
Hb_089140_070 |
0.1061352519 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 3 |
Hb_000098_270 |
0.110140968 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 4 |
Hb_000289_020 |
0.1135730927 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 5 |
Hb_006810_040 |
0.1155721531 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000878_120 |
0.1177712585 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 7 |
Hb_002028_270 |
0.1186240084 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_005701_100 |
0.1238393552 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 9 |
Hb_000390_010 |
0.1271767046 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 10 |
Hb_000270_050 |
0.1284316988 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 11 |
Hb_001971_040 |
0.1284986687 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 12 |
Hb_007126_010 |
0.1286679188 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 13 |
Hb_001377_370 |
0.1298386971 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 14 |
Hb_010080_050 |
0.133931137 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 15 |
Hb_012092_040 |
0.1339732531 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004531_060 |
0.1379097303 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 17 |
Hb_002307_150 |
0.1381311916 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 18 |
Hb_001141_440 |
0.14000218 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 19 |
Hb_006351_030 |
0.1409054115 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MARCH8-like [Jatropha curcas] |
| 20 |
Hb_001950_090 |
0.1409843213 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |