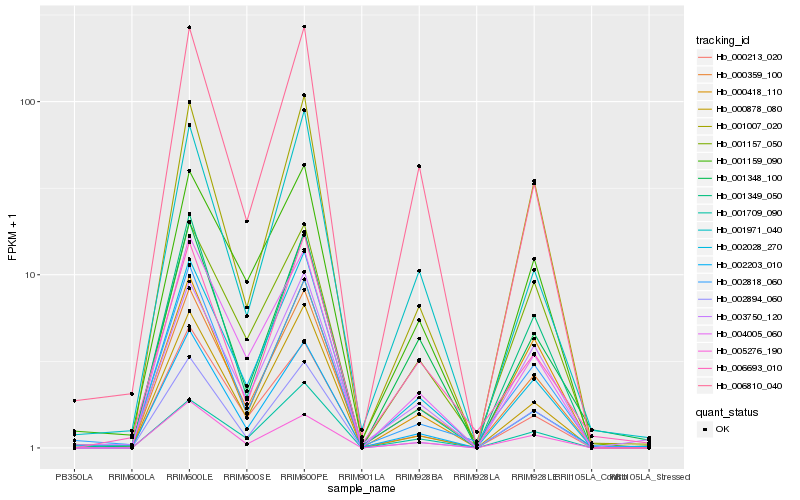
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002203_010 |
0.0730528394 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 3 |
Hb_002028_270 |
0.0822237863 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_004005_060 |
0.0859221991 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 5 |
Hb_002818_060 |
0.0915133431 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_001007_020 |
0.0918607443 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001159_090 |
0.0949526341 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 8 |
Hb_003750_120 |
0.0956799409 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 9 |
Hb_002894_060 |
0.0967879567 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 10 |
Hb_000878_080 |
0.0982064549 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 11 |
Hb_001971_040 |
0.098549929 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 12 |
Hb_005276_190 |
0.099323147 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000418_110 |
0.100241132 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 14 |
Hb_001349_050 |
0.1004653812 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 15 |
Hb_006810_040 |
0.1009186391 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_001709_090 |
0.1016062764 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 17 |
Hb_006693_010 |
0.1017820488 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 18 |
Hb_001157_050 |
0.1030924763 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 19 |
Hb_001348_100 |
0.1034799249 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 20 |
Hb_000213_020 |
0.1043423242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |