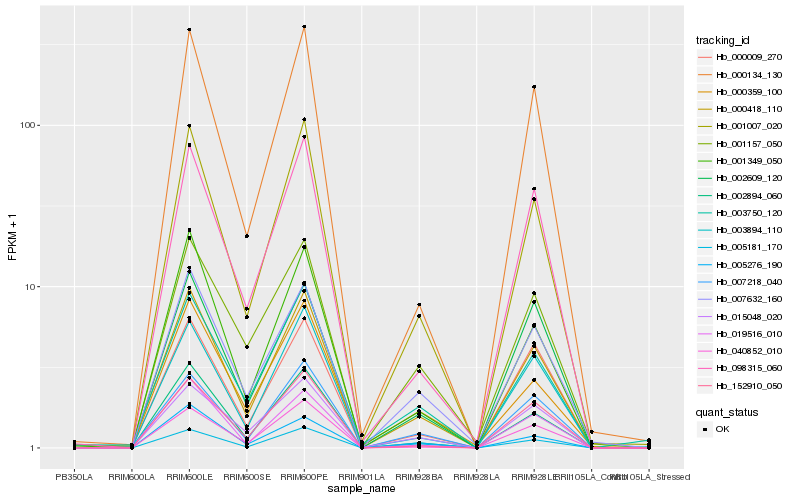
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000418_110 |
0.0 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 2 |
Hb_001007_020 |
0.0463951459 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002894_060 |
0.0583215858 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 4 |
Hb_000134_130 |
0.0784041882 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 5 |
Hb_019516_010 |
0.0794700689 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 6 |
Hb_007632_160 |
0.0801807556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_098315_060 |
0.0820644505 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 8 |
Hb_001349_050 |
0.0844473287 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 9 |
Hb_007218_040 |
0.0846922025 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 10 |
Hb_003894_110 |
0.0877127002 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 11 |
Hb_003750_120 |
0.092708258 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 12 |
Hb_152910_050 |
0.0946962541 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_040852_010 |
0.0953863338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000009_270 |
0.095505099 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 15 |
Hb_015048_020 |
0.0977237158 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |
| 16 |
Hb_000359_100 |
0.100241132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002609_120 |
0.1005751249 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 18 |
Hb_005276_190 |
0.1007521786 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001157_050 |
0.1011083072 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 20 |
Hb_005181_170 |
0.1012041305 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |