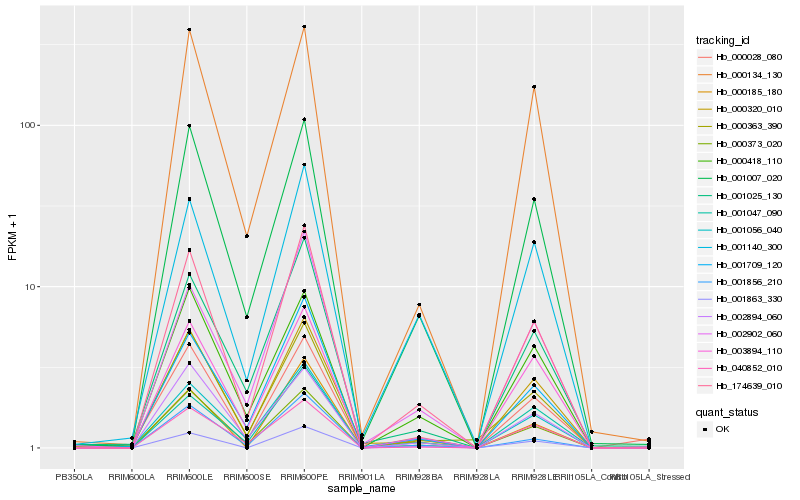
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174639_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_001007_020 |
0.0770151602 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001856_210 |
0.0785799047 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 4 |
Hb_001709_120 |
0.0801661821 |
- |
- |
hypothetical protein RCOM_1313640 [Ricinus communis] |
| 5 |
Hb_001056_040 |
0.0802966552 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 6 |
Hb_001863_330 |
0.0881339543 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000320_010 |
0.0906806813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000185_180 |
0.1017860548 |
- |
- |
hypothetical protein CISIN_1g037968mg, partial [Citrus sinensis] |
| 9 |
Hb_003894_110 |
0.1025530815 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 10 |
Hb_001025_130 |
0.1038070679 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 11 |
Hb_002902_060 |
0.1040692499 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 12 |
Hb_002894_060 |
0.1046651002 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 13 |
Hb_000363_390 |
0.1059342569 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_001140_300 |
0.1065605029 |
- |
- |
PREDICTED: tubulin beta-1 chain [Cicer arietinum] |
| 15 |
Hb_001047_090 |
0.1070384102 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |
| 16 |
Hb_040852_010 |
0.1111296055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000028_080 |
0.1119658818 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 18 |
Hb_000418_110 |
0.1122391924 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 19 |
Hb_000134_130 |
0.1128884529 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 20 |
Hb_000373_020 |
0.115354162 |
- |
- |
PREDICTED: beta-adaptin-like protein A [Jatropha curcas] |