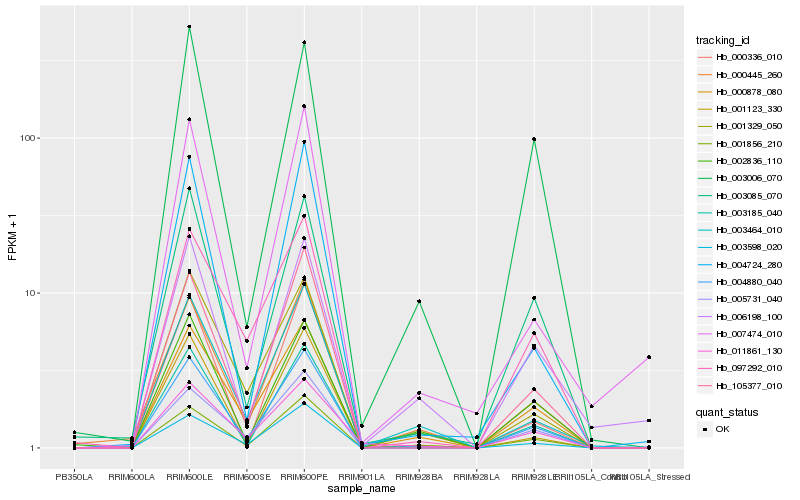
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004880_040 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003185_040 |
0.0667269138 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 3 |
Hb_105377_010 |
0.0776998605 |
- |
- |
hypothetical protein PRUPE_ppa001414mg [Prunus persica] |
| 4 |
Hb_003598_020 |
0.0846199086 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 5 |
Hb_001856_210 |
0.0892860868 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 6 |
Hb_000878_080 |
0.0954158391 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 7 |
Hb_003085_070 |
0.0990210667 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000445_260 |
0.1045442881 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 9 |
Hb_005731_040 |
0.1046301347 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 10 |
Hb_011861_130 |
0.1092248162 |
- |
- |
PREDICTED: uncharacterized protein LOC105634795 [Jatropha curcas] |
| 11 |
Hb_004724_280 |
0.1098770796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003464_010 |
0.1107901814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001123_330 |
0.1122748553 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 14 |
Hb_007474_010 |
0.1132651245 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 15 |
Hb_097292_010 |
0.1159654922 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 16 |
Hb_000336_010 |
0.1187934687 |
transcription factor |
TF Family: C3H |
zinc finger family protein [Populus trichocarpa] |
| 17 |
Hb_003006_070 |
0.1219669103 |
- |
- |
aquaporin [Manihot esculenta] |
| 18 |
Hb_002836_110 |
0.1224801325 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 19 |
Hb_001329_050 |
0.124472254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006198_100 |
0.1262827893 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |