| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
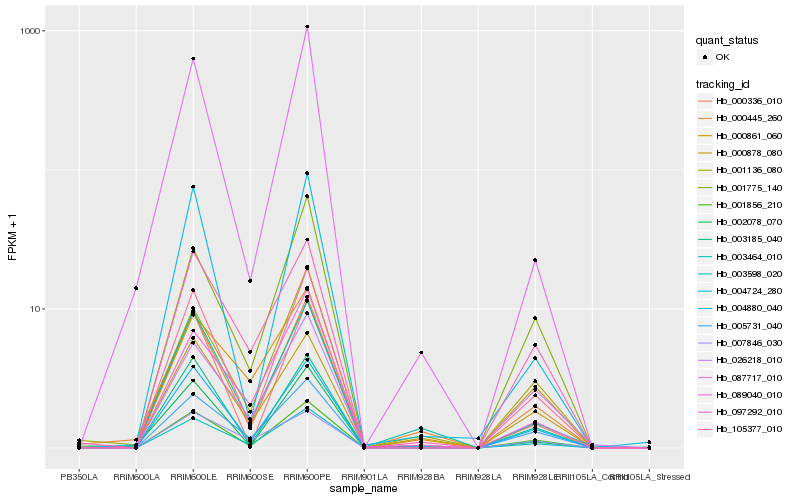
Hb_003598_020 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 2 |
Hb_105377_010 |
0.0495060507 |
- |
- |
hypothetical protein PRUPE_ppa001414mg [Prunus persica] |
| 3 |
Hb_005731_040 |
0.0685749326 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 4 |
Hb_004880_040 |
0.0846199086 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |
| 5 |
Hb_097292_010 |
0.1003705544 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 6 |
Hb_089040_010 |
0.1015732305 |
- |
- |
Glycosyl hydrolase family protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_001856_210 |
0.1128534571 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 8 |
Hb_007846_030 |
0.1132705512 |
- |
- |
Uncharacterized protein TCM_021839 [Theobroma cacao] |
| 9 |
Hb_001775_140 |
0.1135090523 |
- |
- |
Phytosulfokine 3 precursor, putative isoform 2 [Theobroma cacao] |
| 10 |
Hb_001136_080 |
0.1140228566 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 11 |
Hb_003185_040 |
0.1155489987 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 12 |
Hb_000878_080 |
0.1159072137 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 13 |
Hb_000445_260 |
0.1177249496 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 14 |
Hb_003464_010 |
0.1215367905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_026218_010 |
0.1220175632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004724_280 |
0.1245973582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000861_060 |
0.1250343755 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 18 |
Hb_002078_070 |
0.1267816845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000336_010 |
0.1270810332 |
transcription factor |
TF Family: C3H |
zinc finger family protein [Populus trichocarpa] |
| 20 |
Hb_087717_010 |
0.1296350077 |
- |
- |
hypothetical protein JCGZ_01246 [Jatropha curcas] |