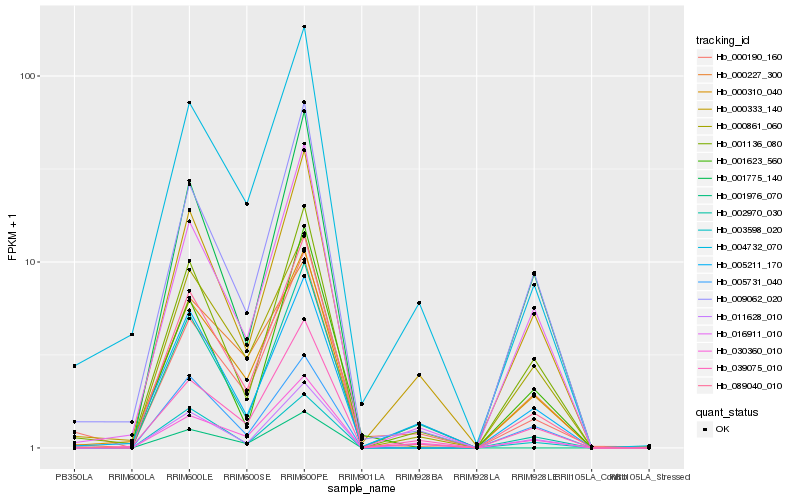
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089040_010 |
0.0 |
- |
- |
Glycosyl hydrolase family protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_003598_020 |
0.1015732305 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 3 |
Hb_002970_030 |
0.1099129332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_016911_010 |
0.1105727025 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 5 |
Hb_001136_080 |
0.1111124179 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 6 |
Hb_039075_010 |
0.1128477389 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 7 |
Hb_000310_040 |
0.1131148314 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 8 |
Hb_000190_160 |
0.1166392025 |
- |
- |
PREDICTED: uncharacterized protein LOC105649940 isoform X2 [Jatropha curcas] |
| 9 |
Hb_009062_020 |
0.1167500293 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 10 |
Hb_011628_010 |
0.1188789007 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 11 |
Hb_001775_140 |
0.1203778484 |
- |
- |
Phytosulfokine 3 precursor, putative isoform 2 [Theobroma cacao] |
| 12 |
Hb_000861_060 |
0.1211864286 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 13 |
Hb_004732_070 |
0.1220530272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001976_070 |
0.124425944 |
- |
- |
PREDICTED: uncharacterized protein LOC101497268 [Cicer arietinum] |
| 15 |
Hb_000333_140 |
0.1268246442 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 16 |
Hb_030360_010 |
0.1275460159 |
- |
- |
- |
| 17 |
Hb_005731_040 |
0.1275682573 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 18 |
Hb_000227_300 |
0.1296375524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001623_560 |
0.1301254986 |
- |
- |
late embryogenesis abundant protein group 9 protein [Arachis hypogaea] |
| 20 |
Hb_005211_170 |
0.1301891204 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |