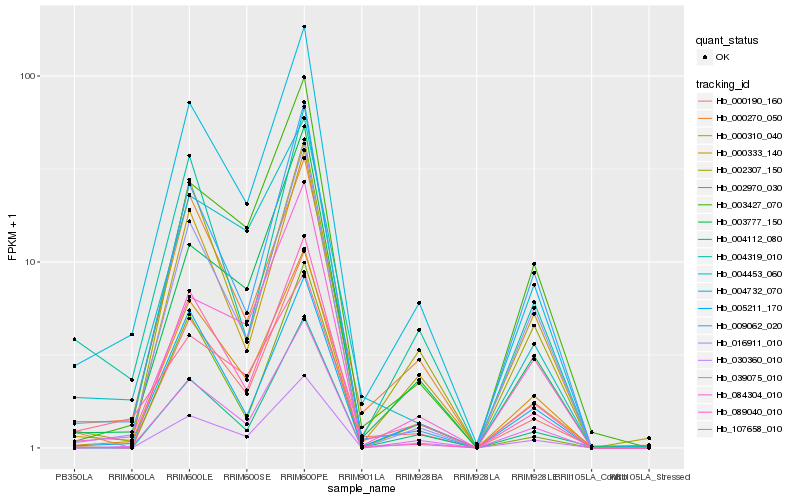
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004732_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000310_040 |
0.0921942921 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 3 |
Hb_000190_160 |
0.0969182542 |
- |
- |
PREDICTED: uncharacterized protein LOC105649940 isoform X2 [Jatropha curcas] |
| 4 |
Hb_039075_010 |
0.1002134672 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 5 |
Hb_003777_150 |
0.1109421283 |
- |
- |
PREDICTED: uncharacterized protein LOC105640938 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000270_050 |
0.1125934948 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 7 |
Hb_030360_010 |
0.1174462983 |
- |
- |
- |
| 8 |
Hb_089040_010 |
0.1220530272 |
- |
- |
Glycosyl hydrolase family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_004453_060 |
0.1245204489 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2 [Jatropha curcas] |
| 10 |
Hb_107658_010 |
0.1266989438 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_002970_030 |
0.1277488191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004112_080 |
0.128258509 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 13 |
Hb_000333_140 |
0.1283285301 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 14 |
Hb_005211_170 |
0.128543552 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 15 |
Hb_003427_070 |
0.1361851675 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 16 |
Hb_002307_150 |
0.1368517573 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 17 |
Hb_084304_010 |
0.1376196131 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 18 |
Hb_016911_010 |
0.1379207578 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 19 |
Hb_004319_010 |
0.1379329121 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 20 |
Hb_009062_020 |
0.1384684739 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |