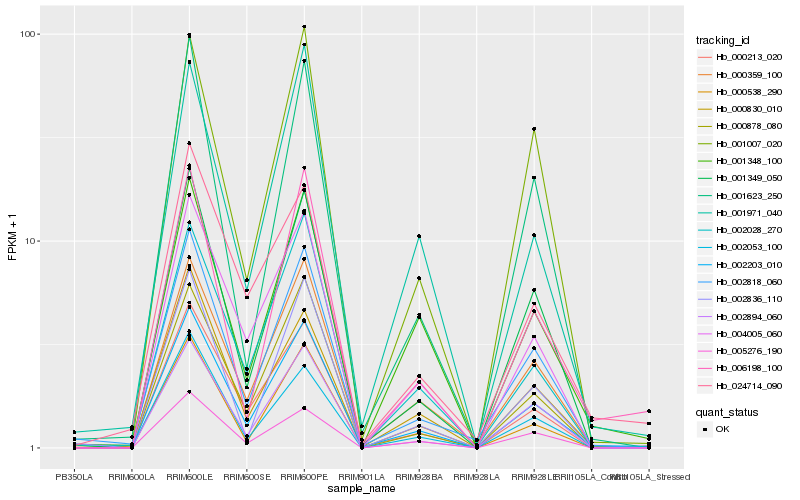
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002203_010 |
0.0 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 2 |
Hb_000213_020 |
0.0685133431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000359_100 |
0.0730528394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001349_050 |
0.0841515107 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 5 |
Hb_004005_060 |
0.0848687001 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 6 |
Hb_002818_060 |
0.0910184005 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_000878_080 |
0.0913310082 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 8 |
Hb_002894_060 |
0.0927197778 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 9 |
Hb_000830_010 |
0.0955894287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002028_270 |
0.0957480154 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 11 |
Hb_005276_190 |
0.0962290469 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002053_100 |
0.1022632644 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 13 |
Hb_000538_290 |
0.1039803282 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 14 |
Hb_006198_100 |
0.1051499559 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 15 |
Hb_001348_100 |
0.1059263862 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 16 |
Hb_001623_250 |
0.1072395465 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 17 |
Hb_001007_020 |
0.1089343825 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001971_040 |
0.1146102539 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 19 |
Hb_002836_110 |
0.1158758064 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 20 |
Hb_024714_090 |
0.1171803645 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |