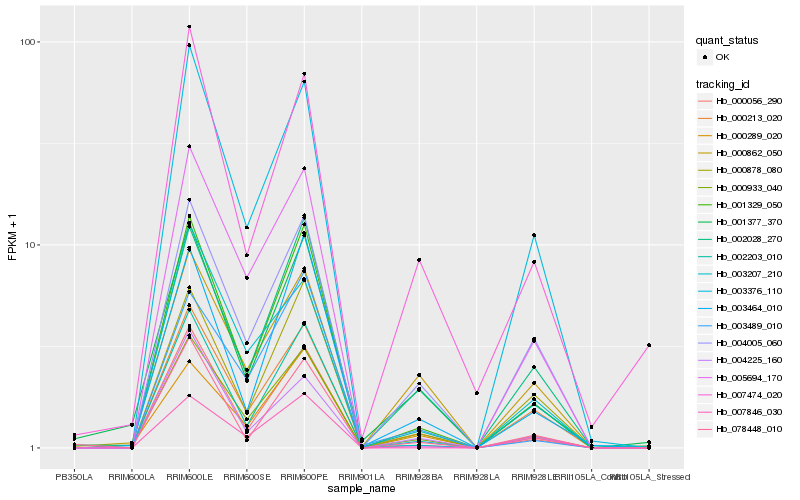
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000933_040 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 2 |
Hb_001329_050 |
0.0639377808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000213_020 |
0.0903006595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000056_290 |
0.0915739185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004005_060 |
0.0991082613 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 6 |
Hb_005694_170 |
0.1080369219 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002028_270 |
0.1120409202 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000289_020 |
0.1158133406 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 9 |
Hb_001377_370 |
0.1192938634 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 10 |
Hb_000878_080 |
0.1193541564 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 11 |
Hb_003489_010 |
0.1194786416 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 12 |
Hb_003376_110 |
0.1225580584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003464_010 |
0.1273454805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004225_160 |
0.1309225833 |
- |
- |
PREDICTED: kiwellin-like [Jatropha curcas] |
| 15 |
Hb_002203_010 |
0.1322022118 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 16 |
Hb_007846_030 |
0.1338988013 |
- |
- |
Uncharacterized protein TCM_021839 [Theobroma cacao] |
| 17 |
Hb_003207_210 |
0.1339814627 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 18 |
Hb_078448_010 |
0.1353493395 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 19 |
Hb_000862_050 |
0.1357649836 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 20 |
Hb_007474_020 |
0.1399306055 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |