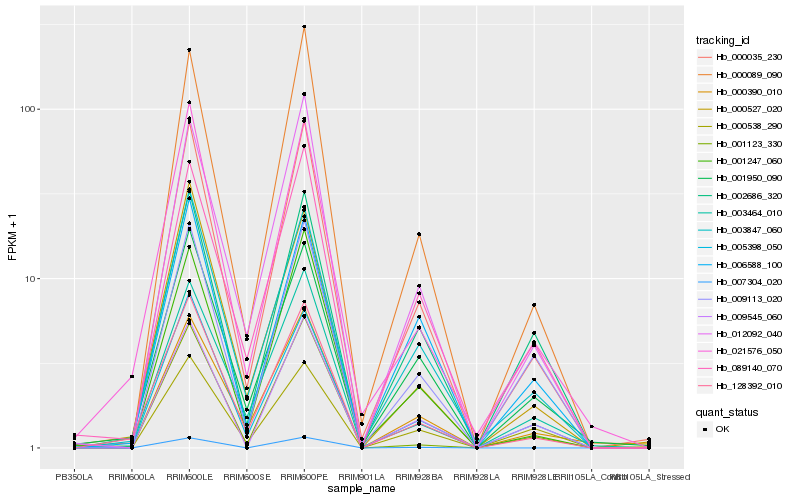
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_230 |
0.0 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 2 |
Hb_001247_060 |
0.0670456441 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000089_090 |
0.0695568345 |
- |
- |
hypothetical protein POPTR_0019s12670g [Populus trichocarpa] |
| 4 |
Hb_003847_060 |
0.0797755789 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 5 |
Hb_012092_040 |
0.0799265697 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005398_050 |
0.0824384321 |
- |
- |
PREDICTED: uncharacterized protein LOC105644215 isoform X1 [Jatropha curcas] |
| 7 |
Hb_128392_010 |
0.0828912821 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009545_060 |
0.0932352463 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |
| 9 |
Hb_021576_050 |
0.0959223957 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 10 |
Hb_003464_010 |
0.0963899441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_089140_070 |
0.0997606298 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 12 |
Hb_000390_010 |
0.1005379705 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 13 |
Hb_000527_020 |
0.1044968116 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 14 |
Hb_009113_020 |
0.1052621385 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 15 |
Hb_001950_090 |
0.1065747471 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 16 |
Hb_001123_330 |
0.108601205 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 17 |
Hb_002686_320 |
0.1116742321 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 18 |
Hb_007304_020 |
0.1121209412 |
- |
- |
PREDICTED: uncharacterized protein LOC100256879 [Vitis vinifera] |
| 19 |
Hb_006588_100 |
0.1167260676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000538_290 |
0.1202266758 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |