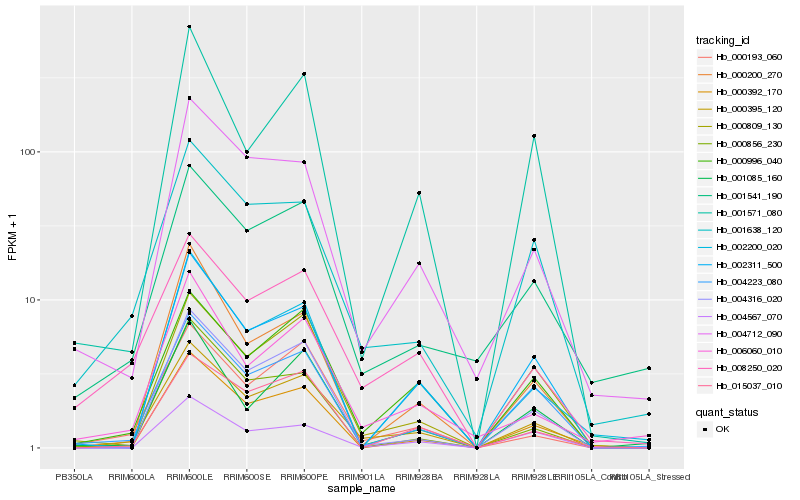
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000395_120 |
0.0 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 2 |
Hb_000996_040 |
0.1187321126 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 3 |
Hb_004223_080 |
0.1264568356 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_004316_020 |
0.1291132421 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 5 |
Hb_000193_060 |
0.1310239765 |
- |
- |
PREDICTED: F-box protein At4g35930 [Jatropha curcas] |
| 6 |
Hb_000809_130 |
0.1322431231 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000392_170 |
0.1324169201 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 8 |
Hb_000200_270 |
0.1360262793 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_015037_010 |
0.1397290719 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 10 |
Hb_001638_120 |
0.1399749524 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 11 |
Hb_004712_090 |
0.1400939314 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_006060_010 |
0.1413727193 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 13 |
Hb_008250_020 |
0.142379825 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002311_500 |
0.1435974113 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 15 |
Hb_001571_080 |
0.1462342456 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 16 |
Hb_004567_070 |
0.1514777268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002200_020 |
0.1520141443 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein RCOM_0137680 [Ricinus communis] |
| 18 |
Hb_001085_160 |
0.1526551029 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 19 |
Hb_000856_230 |
0.1590444382 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 20 |
Hb_001541_190 |
0.1610261033 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |