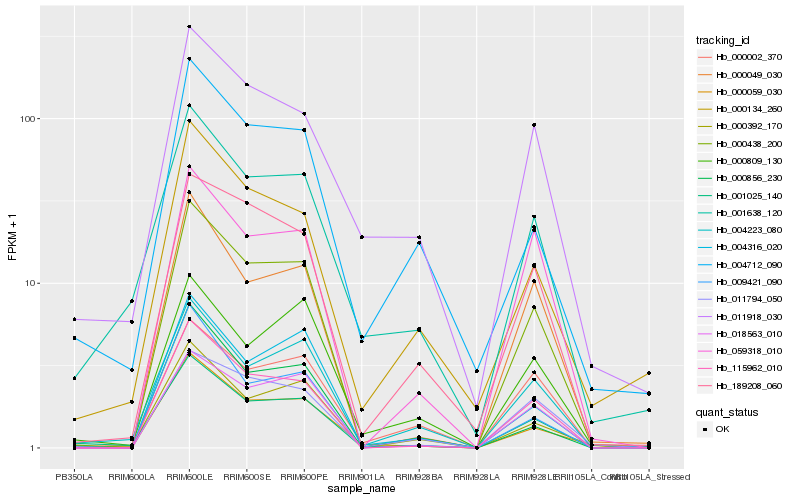
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001025_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s09180g [Populus trichocarpa] |
| 2 |
Hb_004223_080 |
0.1045781703 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_000438_200 |
0.1070131857 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |
| 4 |
Hb_000059_030 |
0.1103619876 |
- |
- |
PREDICTED: chaperone protein dnaJ 10 [Jatropha curcas] |
| 5 |
Hb_115962_010 |
0.1122158985 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 6 |
Hb_001638_120 |
0.1213413715 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 7 |
Hb_000049_030 |
0.1242779958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000392_170 |
0.1282217848 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 9 |
Hb_018563_010 |
0.1397613266 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 10 |
Hb_011918_030 |
0.1430603606 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 11 |
Hb_004316_020 |
0.1467591211 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 12 |
Hb_009421_090 |
0.1472675168 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 13 |
Hb_000809_130 |
0.1476135111 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000856_230 |
0.1494985021 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 15 |
Hb_000134_260 |
0.1510116886 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000002_370 |
0.1518767136 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 17 |
Hb_004712_090 |
0.1528152096 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 18 |
Hb_059318_010 |
0.1528900095 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 19 |
Hb_189208_060 |
0.1530645292 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 20 |
Hb_011794_050 |
0.1537131089 |
- |
- |
transcription factor, putative [Ricinus communis] |