| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
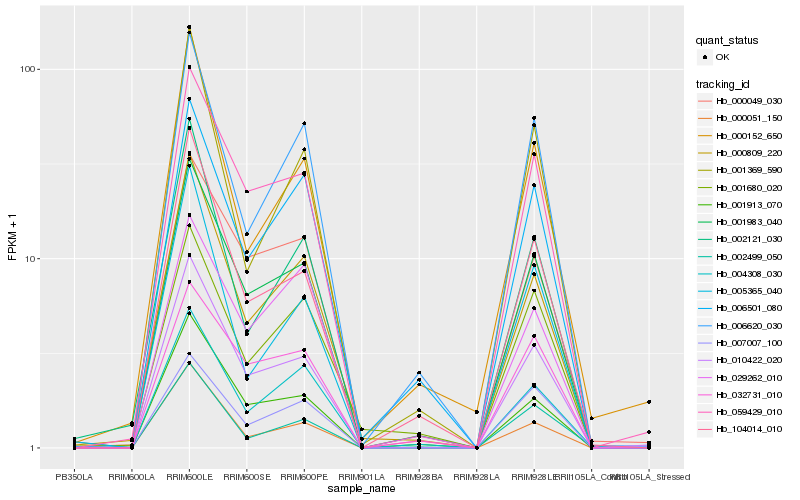
Hb_001983_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 2 |
Hb_010422_020 |
0.0462496545 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 3 |
Hb_059429_010 |
0.0465311102 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 4 |
Hb_001913_070 |
0.0827304364 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 5 |
Hb_000049_030 |
0.0903582051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000809_220 |
0.0908965918 |
- |
- |
PREDICTED: 3-ketodihydrosphingosine reductase [Jatropha curcas] |
| 7 |
Hb_000051_150 |
0.0959454215 |
- |
- |
hypothetical protein CICLE_v10003316mg [Citrus clementina] |
| 8 |
Hb_104014_010 |
0.1028718258 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004308_030 |
0.1032420192 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 10 |
Hb_006501_080 |
0.1082161707 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 11 |
Hb_006620_030 |
0.1091388182 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 12 |
Hb_032731_010 |
0.114044893 |
- |
- |
RING-H2 finger protein ATL5M precursor, putative [Ricinus communis] |
| 13 |
Hb_007007_100 |
0.1197525249 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 14 |
Hb_029262_010 |
0.1234367549 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 15 |
Hb_001680_020 |
0.1261730151 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 16 |
Hb_002121_030 |
0.1263625164 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 17 |
Hb_002499_050 |
0.1298693394 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_001369_590 |
0.1306437149 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 19 |
Hb_005365_040 |
0.1328525061 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 20 |
Hb_000152_650 |
0.1346938939 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |