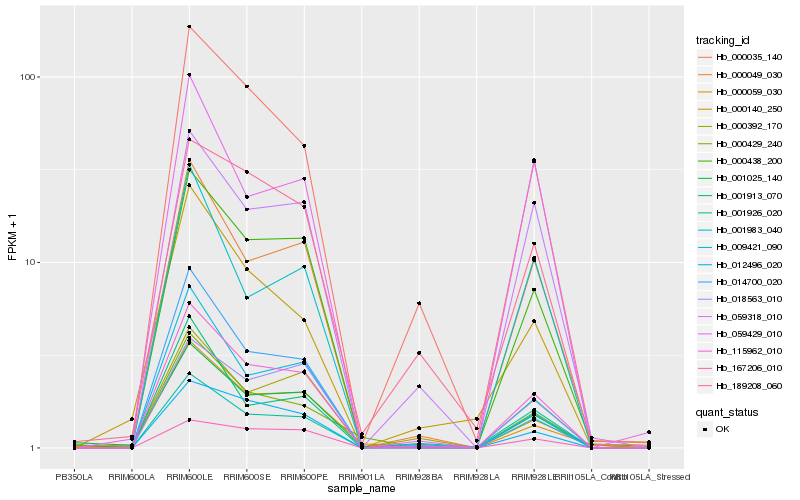
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115962_010 |
0.0 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 2 |
Hb_000438_200 |
0.0610293863 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |
| 3 |
Hb_000049_030 |
0.0865478275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000035_140 |
0.0947075849 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 5 |
Hb_012496_020 |
0.1062100409 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 6 |
Hb_009421_090 |
0.1100327805 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 7 |
Hb_001025_140 |
0.1122158985 |
- |
- |
hypothetical protein POPTR_0015s09180g [Populus trichocarpa] |
| 8 |
Hb_059318_010 |
0.1187748427 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 9 |
Hb_018563_010 |
0.1205824243 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 10 |
Hb_001913_070 |
0.1209345122 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 11 |
Hb_014700_020 |
0.1286888792 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 12 |
Hb_000429_240 |
0.128748 |
- |
- |
PREDICTED: uncharacterized protein LOC105645185 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000059_030 |
0.1293365259 |
- |
- |
PREDICTED: chaperone protein dnaJ 10 [Jatropha curcas] |
| 14 |
Hb_167206_010 |
0.1323283743 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 15 |
Hb_059429_010 |
0.1335762946 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 16 |
Hb_001926_020 |
0.1370680787 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 17 |
Hb_000140_250 |
0.1399870246 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 18 |
Hb_189208_060 |
0.14216472 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 19 |
Hb_001983_040 |
0.1422992195 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 20 |
Hb_000392_170 |
0.1439134574 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |