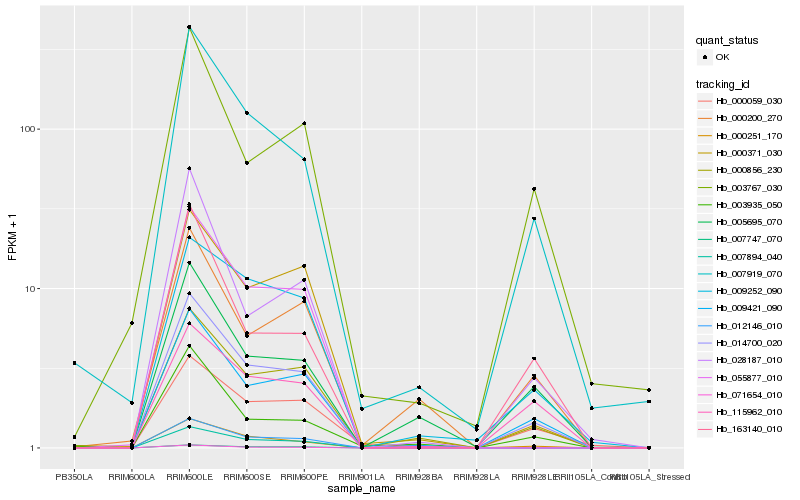
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014700_020 |
0.0 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 2 |
Hb_009421_090 |
0.0654632272 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 3 |
Hb_000251_170 |
0.0723973692 |
- |
- |
leucine-rich repeat transmembrane protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000856_230 |
0.094933673 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 5 |
Hb_007919_070 |
0.0996202686 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 6 |
Hb_115962_010 |
0.1286888792 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 7 |
Hb_000371_030 |
0.1291110205 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: anthocyanidin 3-O-glucosyltransferase 5 [Prunus mume] |
| 8 |
Hb_009252_090 |
0.1313643277 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_071654_010 |
0.1347104744 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 10 |
Hb_012146_010 |
0.1361304499 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 11 |
Hb_055877_010 |
0.1372232569 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_005695_070 |
0.1374103734 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 13 |
Hb_163140_010 |
0.1405812899 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 14 |
Hb_007894_040 |
0.1406143904 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_028187_010 |
0.1412205301 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 16 |
Hb_003935_050 |
0.1431844212 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_000059_030 |
0.1433892523 |
- |
- |
PREDICTED: chaperone protein dnaJ 10 [Jatropha curcas] |
| 18 |
Hb_003767_030 |
0.1439685845 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 19 |
Hb_000200_270 |
0.1469484986 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_007747_070 |
0.1472474918 |
- |
- |
PREDICTED: casparian strip membrane protein 1-like [Jatropha curcas] |