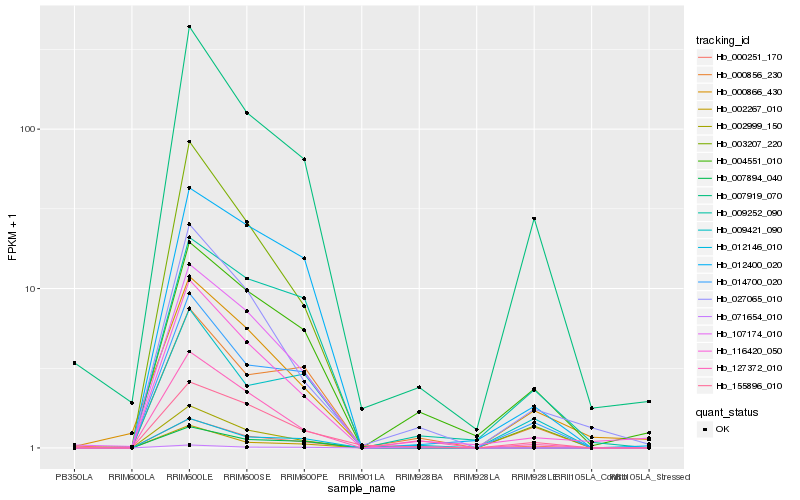
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000251_170 |
0.0 |
- |
- |
leucine-rich repeat transmembrane protein kinase, putative [Ricinus communis] |
| 2 |
Hb_014700_020 |
0.0723973692 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 3 |
Hb_007919_070 |
0.087763623 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 4 |
Hb_002999_150 |
0.1295986821 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 5 |
Hb_155896_010 |
0.1297408023 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 6 |
Hb_003207_220 |
0.1302016641 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 7 |
Hb_009421_090 |
0.1323098733 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 8 |
Hb_071654_010 |
0.1376296734 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 9 |
Hb_012400_020 |
0.1380755918 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 10 |
Hb_000866_430 |
0.1417292988 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 11 |
Hb_007894_040 |
0.142045996 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_116420_050 |
0.1426607255 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 13 |
Hb_012146_010 |
0.142781561 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 14 |
Hb_009252_090 |
0.1428561204 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_107174_010 |
0.1445365209 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_004551_010 |
0.1454529271 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 17 |
Hb_127372_010 |
0.1456834139 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 18 |
Hb_000856_230 |
0.1469776139 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 19 |
Hb_002267_010 |
0.1475597296 |
- |
- |
hypothetical protein SORBIDRAFT_01g040370 [Sorghum bicolor] |
| 20 |
Hb_027065_010 |
0.14959721 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |