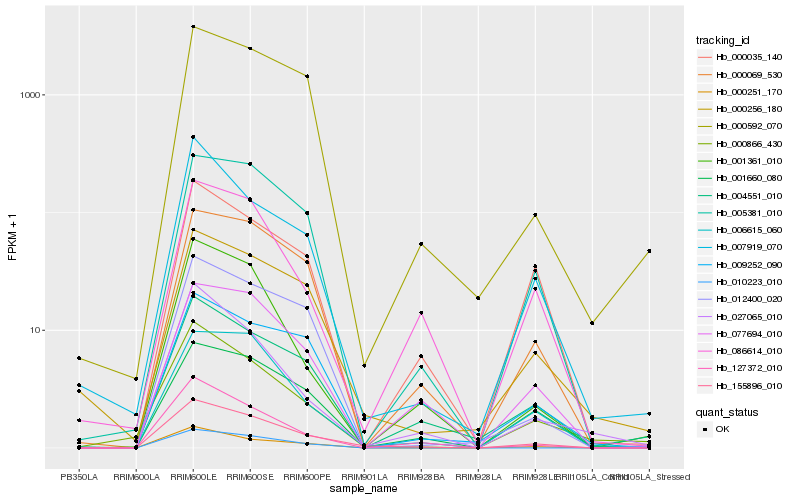
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155896_010 |
0.0 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 2 |
Hb_000251_170 |
0.1297408023 |
- |
- |
leucine-rich repeat transmembrane protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000069_530 |
0.1331682522 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 4 |
Hb_005381_010 |
0.1332743109 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006615_060 |
0.1399725913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_077694_010 |
0.1419413005 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |
| 7 |
Hb_127372_010 |
0.1422210167 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 8 |
Hb_004551_010 |
0.1445442282 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 9 |
Hb_001660_080 |
0.1453173957 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 10 |
Hb_012400_020 |
0.1485626754 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 11 |
Hb_000256_180 |
0.1490318959 |
- |
- |
PREDICTED: uncharacterized protein LOC105637589 [Jatropha curcas] |
| 12 |
Hb_001361_010 |
0.1524690563 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 13 |
Hb_009252_090 |
0.152597023 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_007919_070 |
0.1533894247 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 15 |
Hb_000592_070 |
0.1561770518 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 16 |
Hb_010223_010 |
0.1575386973 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 17 |
Hb_086614_010 |
0.1581602712 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000035_140 |
0.1583480081 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 19 |
Hb_000866_430 |
0.1585620799 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 20 |
Hb_027065_010 |
0.1586034148 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |