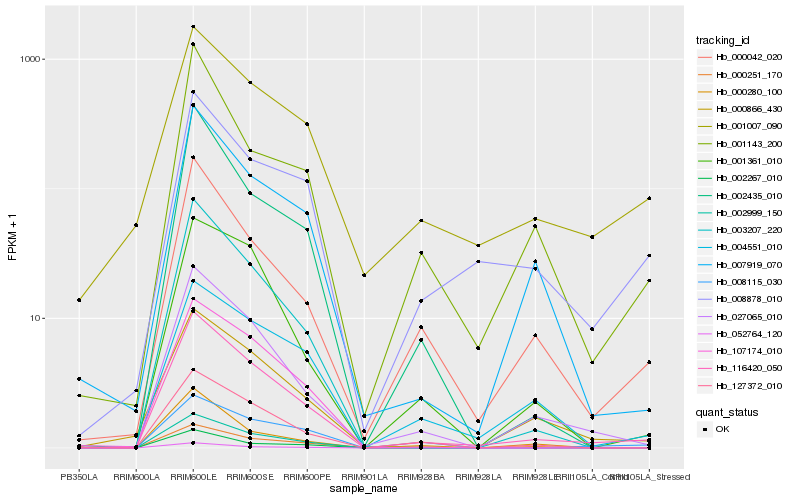
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116420_050 |
0.0 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 2 |
Hb_027065_010 |
0.094988587 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 3 |
Hb_127372_010 |
0.1214255637 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 4 |
Hb_003207_220 |
0.1271554595 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 5 |
Hb_000042_020 |
0.129609294 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_007919_070 |
0.1307519422 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 7 |
Hb_000866_430 |
0.1368007559 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 8 |
Hb_002999_150 |
0.1380573336 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 9 |
Hb_000251_170 |
0.1426607255 |
- |
- |
leucine-rich repeat transmembrane protein kinase, putative [Ricinus communis] |
| 10 |
Hb_002435_010 |
0.1442621884 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 11 |
Hb_107174_010 |
0.1454384266 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001361_010 |
0.1495893286 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 13 |
Hb_004551_010 |
0.1507155281 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 14 |
Hb_001143_200 |
0.1543046472 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 15 |
Hb_052764_120 |
0.1580301493 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 16 |
Hb_008878_010 |
0.1592921873 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 17 |
Hb_001007_090 |
0.1594135434 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 18 |
Hb_000280_100 |
0.1653749953 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 19 |
Hb_002267_010 |
0.1658493423 |
- |
- |
hypothetical protein SORBIDRAFT_01g040370 [Sorghum bicolor] |
| 20 |
Hb_008115_030 |
0.168546888 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |