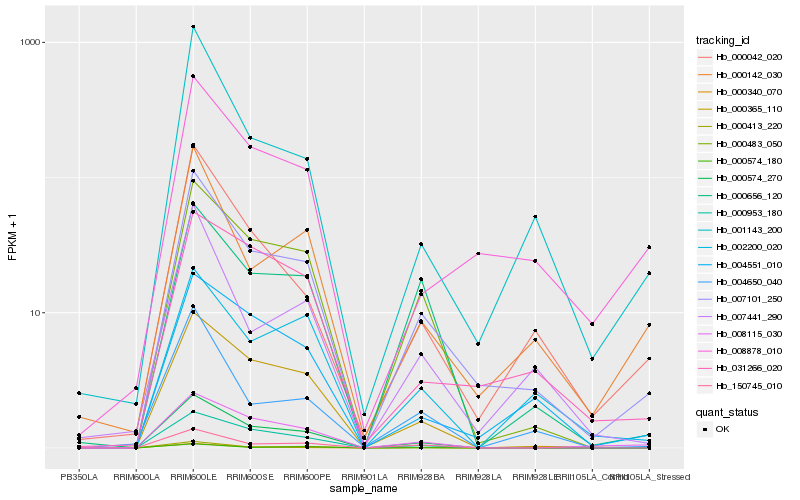
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007101_250 |
0.0 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 2 |
Hb_000574_270 |
0.1298069596 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 3 |
Hb_000483_050 |
0.1344129533 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 4 |
Hb_000365_110 |
0.13823222 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 5 |
Hb_150745_010 |
0.1457096512 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 6 |
Hb_000142_030 |
0.1471032601 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_008878_010 |
0.1503497893 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 8 |
Hb_001143_200 |
0.1521013314 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 9 |
Hb_004551_010 |
0.1534052411 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 10 |
Hb_000042_020 |
0.1534181437 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_008115_030 |
0.1554077607 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000413_220 |
0.1574371757 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 13 |
Hb_007441_290 |
0.1633889719 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 14 |
Hb_000340_070 |
0.1637461216 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 15 |
Hb_000953_180 |
0.1644635663 |
- |
- |
- |
| 16 |
Hb_000656_120 |
0.1646500773 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 17 |
Hb_004650_040 |
0.1647295635 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 18 |
Hb_000574_180 |
0.1663647932 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 19 |
Hb_031266_020 |
0.167690646 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002200_020 |
0.1692593134 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein RCOM_0137680 [Ricinus communis] |