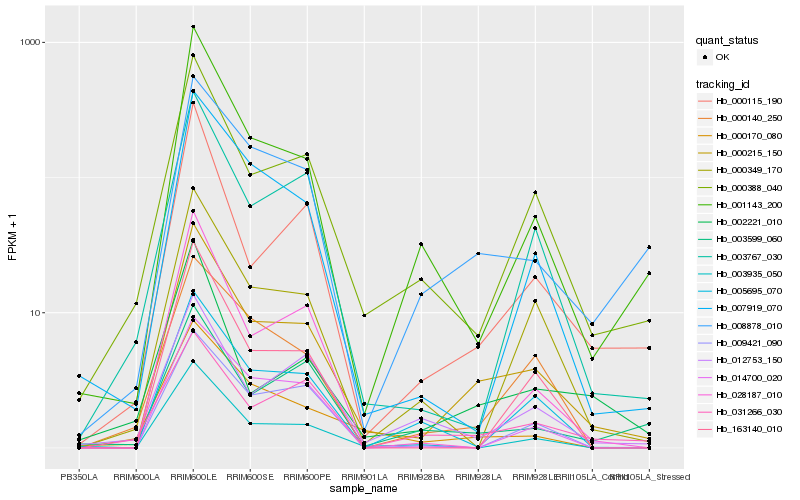
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100249186 [Vitis vinifera] |
| 2 |
Hb_000388_040 |
0.1344366919 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 3 |
Hb_000115_190 |
0.1505424439 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 4 |
Hb_001143_200 |
0.1532085379 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 5 |
Hb_008878_010 |
0.1546177265 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 6 |
Hb_003767_030 |
0.1561284356 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 7 |
Hb_007919_070 |
0.1580955503 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 8 |
Hb_003935_050 |
0.1613079455 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000349_170 |
0.1649166075 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 10 |
Hb_163140_010 |
0.168451099 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 11 |
Hb_000170_080 |
0.169289 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 12 |
Hb_028187_010 |
0.17133073 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 13 |
Hb_005695_070 |
0.1761462701 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 14 |
Hb_000140_250 |
0.1781732384 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 15 |
Hb_012753_150 |
0.1789907041 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 16 |
Hb_014700_020 |
0.179019613 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 17 |
Hb_031266_030 |
0.1800873315 |
- |
- |
Kinase superfamily protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_009421_090 |
0.1806842109 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 19 |
Hb_002221_010 |
0.1826364009 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3-like [Jatropha curcas] |
| 20 |
Hb_003599_060 |
0.1827638315 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |