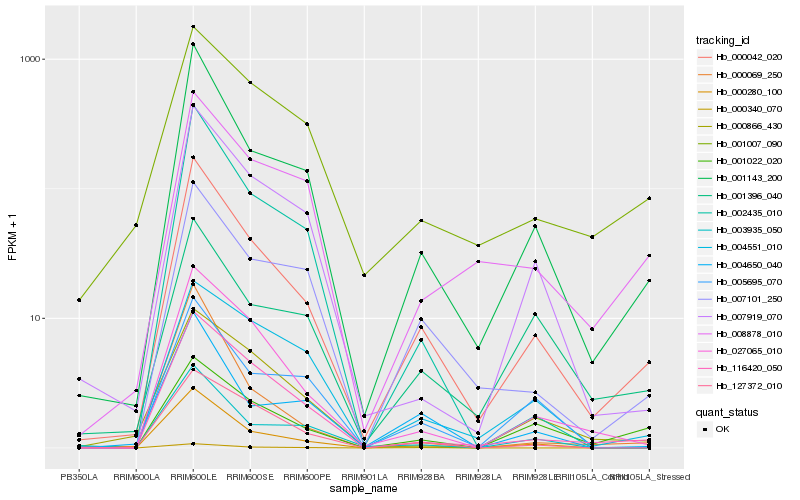
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_020 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_001143_200 |
0.0884574335 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 3 |
Hb_000280_100 |
0.1196191962 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 4 |
Hb_116420_050 |
0.129609294 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 5 |
Hb_027065_010 |
0.1361413682 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 6 |
Hb_007101_250 |
0.1534181437 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 7 |
Hb_127372_010 |
0.1565309635 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 8 |
Hb_007919_070 |
0.1580518004 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 9 |
Hb_004650_040 |
0.1669474878 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 10 |
Hb_000340_070 |
0.1680581094 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 11 |
Hb_002435_010 |
0.1683136253 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 12 |
Hb_005695_070 |
0.1706347704 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 13 |
Hb_003935_050 |
0.1707465387 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_008878_010 |
0.1724400224 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 15 |
Hb_001007_090 |
0.1750517644 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 16 |
Hb_001022_020 |
0.1758728767 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 17 |
Hb_000069_250 |
0.1827282732 |
- |
- |
hypothetical protein JCGZ_14250 [Jatropha curcas] |
| 18 |
Hb_001396_040 |
0.1833129665 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 19 |
Hb_004551_010 |
0.184198402 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 20 |
Hb_000866_430 |
0.1844634532 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |