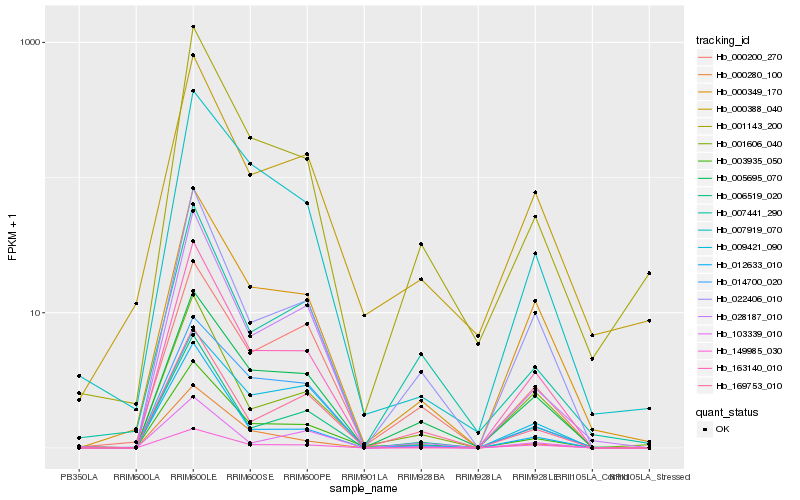
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003935_050 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_001143_200 |
0.1187008266 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 3 |
Hb_005695_070 |
0.1199444563 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 4 |
Hb_163140_010 |
0.1205498466 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 5 |
Hb_007919_070 |
0.1239045235 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 6 |
Hb_000280_100 |
0.1244349653 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 7 |
Hb_028187_010 |
0.1263126229 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 8 |
Hb_149985_030 |
0.1266124149 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 9 |
Hb_006519_020 |
0.128394773 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 10 |
Hb_022406_010 |
0.1312533022 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000349_170 |
0.1330221494 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 12 |
Hb_012633_010 |
0.1351782809 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 13 |
Hb_169753_010 |
0.1358693236 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 14 |
Hb_000388_040 |
0.1375817626 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 15 |
Hb_007441_290 |
0.1419902418 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 16 |
Hb_014700_020 |
0.1431844212 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 17 |
Hb_009421_090 |
0.1431872526 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 18 |
Hb_000200_270 |
0.1443942758 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001606_040 |
0.1462908794 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 20 |
Hb_103339_010 |
0.1465545421 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |