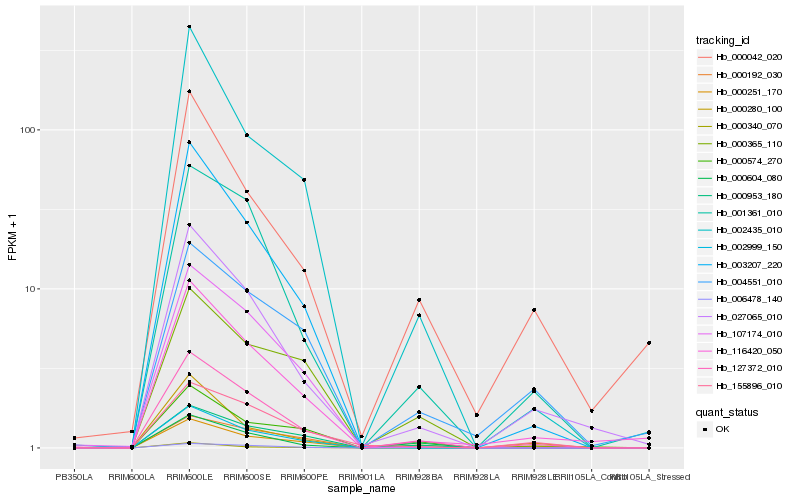
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127372_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 2 |
Hb_001361_010 |
0.0983140524 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 3 |
Hb_027065_010 |
0.1037068359 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 4 |
Hb_116420_050 |
0.1214255637 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 5 |
Hb_000953_180 |
0.126403948 |
- |
- |
- |
| 6 |
Hb_003207_220 |
0.1290639407 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 7 |
Hb_000604_080 |
0.1320211361 |
- |
- |
Fgenesh protein [Medicago truncatula] |
| 8 |
Hb_107174_010 |
0.13892901 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_002999_150 |
0.1401108465 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 10 |
Hb_006478_140 |
0.1407337357 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 11 |
Hb_002435_010 |
0.141526133 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 12 |
Hb_000192_030 |
0.1420905533 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 13 |
Hb_155896_010 |
0.1422210167 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 14 |
Hb_000251_170 |
0.1456834139 |
- |
- |
leucine-rich repeat transmembrane protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000574_270 |
0.1471892451 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 16 |
Hb_000280_100 |
0.1490771274 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 17 |
Hb_000365_110 |
0.1537693501 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 18 |
Hb_000042_020 |
0.1565309635 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_004551_010 |
0.1583222735 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 20 |
Hb_000340_070 |
0.1596849702 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |